We caught up with authors from some of the journal’s first papers to discuss their work, then and now.
To celebrate ten years of Open Biology, we catch up with some our first authors and reflect on their research and experience of publishing with the journal.
Mariann Bienz, MRC Laboratory of Molecular Biology
What was the history behind the work that you published in Open Biology? Did you face any challenges in the project?
It had been known for more than 20 years that the great majority of colorectal cancers are linked to inactivating mutations in APC. It subsequently turned out that the APC tumour suppressor functions to oppose Wnt signalling, an ancient cell communication pathway conserved in all animals and humans to specify cell fates during development and tissue homeostasis. Work in many labs had established that APC binds to a protein called Axin to form a complex that promotes the destruction of the Wnt effector beta-catenin. How APC enables Axin to do this was unclear.
that the great majority of colorectal cancers are linked to inactivating mutations in APC. It subsequently turned out that the APC tumour suppressor functions to oppose Wnt signalling, an ancient cell communication pathway conserved in all animals and humans to specify cell fates during development and tissue homeostasis. Work in many labs had established that APC binds to a protein called Axin to form a complex that promotes the destruction of the Wnt effector beta-catenin. How APC enables Axin to do this was unclear.
Axin complexes can be visualised as discrete puncta in embryonic and larval tissues of Drosophila. In our study, we used this animal model to tackle this question. We thus discovered that APC mutant cells do not show Axin puncta, which implicated APC in the assembly of the beta-catenin destruction complex (also named Axin degradasome). Since we made similar observations in APC mutant human colorectal cancer cells, this indicated that we had uncovered a conserved function of APC in promoting Axin complex assembly.
How has your group’s work changed since your paper was published?
We knew at the time that Axin complex assembly also relies on Axin's ability to undergo dynamic head-to-tail polymerisation through its DIX domain. DIX domains have been a main focus of our work for many years: they are found in many proteins and, as we discovered, provide an elegantly parsimonious mechanism to assemble multivalent complexes (also known as biomolecular condensates) with high binding avidity for effectors. But DIX domains assemble two-dimensional filaments, whereas Axin complexes are three-dimensional structures, which puzzled us for quite a while. Evidently, the two-dimensional DIX filaments needed to be meshed into three-dimensional molecular assemblies by mutual cross-linking. A few years later, we came across one mechanism of cross-linking while studying Dishevelled, a Wnt signalling protein which polymerises through its DIX domain to assemble multivalent complexes called Wnt signalosomes. In these complexes, cross-links are provided by a dimerising domain present in Dishevelled itself. However, Axin does not contain a dimerising domain, but we realised that its cross-linking into three-dimensional assemblies could be provided by its binding partner APC which can homodimerise. The Axin complex therefore uses an indirect cross-linking system in which the cross-linking of dynamic two-dimensional filaments is provided by a homodimerising binding partner.
The Dishevelled paradigm: cross-linking of short dynamic polymers.
Stephen Baker, University of Cambridge/Wellcome Africa- Asia programme
What first attracted you to working in microbiology and infectious diseases?
I started working assembling genomes in 1998 and the first one I worked on was Salmonella typhi, which causes typhoid fever. It was this organism I picked up as a focus of my research and I am still working on it today. I had no formal training in microbiology but found the interplay between genetics/genomics and the ability of an organism to cause disease really interesting. I also found some of the more basic research less interesting so tried to find things that were more applied, so using genomics to understand how bacteria spread, evolve, become resistant to antibiotics (and what we can do about it) became a natural path forward.
and the first one I worked on was Salmonella typhi, which causes typhoid fever. It was this organism I picked up as a focus of my research and I am still working on it today. I had no formal training in microbiology but found the interplay between genetics/genomics and the ability of an organism to cause disease really interesting. I also found some of the more basic research less interesting so tried to find things that were more applied, so using genomics to understand how bacteria spread, evolve, become resistant to antibiotics (and what we can do about it) became a natural path forward.
What was the history behind the work that you published in Open Biology? Did you face any challenges in the project?
There was a lot of history with this paper. It seems old fashioned now, but at the time it was a new way to link genotyping data with epidemiology. I remember it being an enormous amount of work for all involved, but the data that were generated were really exciting and the work was a change in the direction for bacterial genotyping papers. It got largely ignored for a period, but slowly began to get picked up as people became more familiar with the work. It really was a starting point for the work I would perform for the next decade in Asia and of all my papers it’s the one I get asked about the most and is probably still my favourite.
Petros Ligoxygakis, University of Oxford
What first attracted you to working in innate immunity?
Maybe not surprisingly, I remember exactly the moment that this happened. I was in the middle of my PhD working at the Institute of Molecular Biology and Biotechnology in Crete (Greece) and someone gave me the landmark 1996 Cell paper by Lemaitre et al to read. This paper showed that Toll/NF-κB signalling was important for the Drosophila anti-fungal response and single-handedly resurrected innate immunity as a field. I was blown away. It seemed that there was so much to learn here; even the fact that insects had an immune system, components of which proved later to be evolutionary conserved from sea urchins to humans, seemed monumental. Of course, that paper is now Nobel Prize material but we wouldn’t know this until 2011. After I finished my PhD I had the good fortune to work as a postdoc in the Hoffmann lab between 2000 and 2004.
What was the history behind the Review that you published in Open Biology? Do you recall facing any challenges in the project?
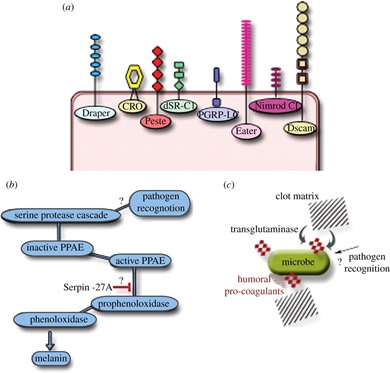
The history behind the review was the fact that an article summarising all different aspects of Drosophila immunity beyond the Toll/NF-κB signalling that everyone was covering those days was lacking. Also, I wanted to cover areas where future work could go. The seeds of our subsequent work with trypanosomatid parasites in Drosophila was planted as a coda to the open questions at the end of the article. The Royal Society was a big pull factor for me to want to publish such a review in their then new journal and I feel that it was really well-received. I think an updated version is due…The field is certainly as vibrant as it has ever been!
Layers of Drosophila immunity.
Tomáš Pluskal, IOCB Prague
When you published your article you were at OIST; how was your time there?
I spent ten years at OIST, from 2005 to 2015. During those years, I witnessed the growth of OIST from a small-scale research lab located in a temporary building into the massive graduate university with a spaceship-like, futuristic campus and first-class research facilities that few other universities can match. In a relatively short time, OIST has become a well-recognized, international research institution of global significance. I really enjoyed my time in Okinawa, it is a beautiful island with spectacular coral reefs and nature, and also very hospitable people who maintain their culture very well.
During those years, I witnessed the growth of OIST from a small-scale research lab located in a temporary building into the massive graduate university with a spaceship-like, futuristic campus and first-class research facilities that few other universities can match. In a relatively short time, OIST has become a well-recognized, international research institution of global significance. I really enjoyed my time in Okinawa, it is a beautiful island with spectacular coral reefs and nature, and also very hospitable people who maintain their culture very well.
Now at IOCB Prague, did you find it at all daunting starting your own group?
In the beginning, I was worried if I could attract good students and postdocs, because the Czech Academy of Sciences does not have such a strong reputation like MIT or the Max Planck Institute. In fact, that turned out to be completely false, I was able to recruit a group of super talented and motivated people, and the research at my lab is now running at full speed. Of course, being a junior PI is very busy and sometimes stressful, especially getting funding. But the IOCB is very welcoming and tries really hard to make the life of junior PIs as easy as possible, with excellent administrative and technical support.
Miratul Muqit, University of Dundee
Do you remember what you were doing when the paper was finally published? What were you and your group focused on at the time?
We had spent three years trying to develop a system to assess PINK1 catalytic activity. Once we achieved this through the discovery of active insect orthologues published in our first paper, we were focused on exploiting this discovery to screen and identify the elusive PINK1 substrate. At the time of this publication, we had observed that PINK1 robustly phosphorylated the Parkin ubiquitin E3 ligase at the conserved Ser65 residue thereby providing a direct mechanistic link of how these two proteins interact. We were working full speed to publish these findings which subsequently appeared in Open Biology the following year.
What has been the highlight of your research career so far?
The discovery that PINK1 phosphorylated Parkin at Serine65 which catalysed a flurry of research activity in the field to define how these two Parkinson’s proteins orchestrate the removal of damaged mitochondria by mitophagy. We had been working intensely on the problem for four years with few breaks and so when we obtained the key results from experiments leading up to the discovery, it was an extremely exciting time in the lab.

Miratul Muqit's Research Group.
Open Biology celebrates ten years as the Royal Society’s open access journal for cellular and molecular biology. Find out more about becoming an author (PDF) and how to submit.
Picture captions and credits:
1.) Photo by National Cancer Institute on Unsplash. Brain Cancer Chromosomes. Created by Thomas Ried, 2014
2.) Stephen Baker: own portrait
3.) Mariann Bienz: own portrait
4.) The Dishevelled paradigm: cross-linking of short dynamic polymers. Credit Mariann Bienz
5.) Layers of Drosophila immunity, Open Biology doi.org/10.1098/rsob.120075
6.) Tomáš Pluskal: own portrait
7.) Miratul Muqit's Research Group. Credit Miratul Muqit