We catch up with authors of some of the journal’s first papers to discuss their work, then and now.
To celebrate ten years of Open Biology, we catch up with some our first authors and reflect on their research and experience of publishing with the journal. In parts one and two we shared the stories of some of our authors from around the world, including the author of our very first published article.
Elizabeth Skippington, Genentech and Mark A. Ragan, The University of Queensland
What were you doing when the paper was published? What was your group focused on at the time?
[Mark]: In 2012, my research group at the Institute for Molecular Biosciences – postdocs, PhD students and trainees – combined core skills in comparative, computational and evolutionary biology, and network inference. We worked at multi-genome scale and liked problems that could benefit from scalable algorithms and high-performance computing. At least, our funders liked those problems; I can’t speak for my students and postdocs.
Bacterial genomics was a rich source of such problems and had been an area of focus for my group since I moved to Australia in 2000. We also had funding in cancer genomics, and dabbled in crop plant and soil metagenomics, but bacterial evolutionary genomics was our bread and butter. The diversity of bacterial (and archaeal) genomes has come about not only by classical descent with modification, but also by lateral genetic transfer (LGT) mediated by viruses, plasmids and/or direct uptake of DNA from the environment. Senior postdocs in our group managed large, well-annotated datasets of genome sequences, aligned orthogroups of genes and proteins, and phylogenetic trees. We had algorithms and workflows to infer recombination breakpoints, and study the evolutionary dynamics of gene fragments – not just whole genes. We had taken early steps toward alignment-free phylogenetics, but hadn’t yet invested heavily in k-mer approaches.
After examining LGT broadly across bacteria and archaea, it was time to focus on evolutionary dynamics within specific families or genera. Data were becoming available, and Elizabeth led our move into this finer-scale work. For her thesis project, she focused on genomes of Escherichia coli and Shigella, identifying lateral regions and studying the impact of LGT on transcriptional regulation (BMC Genomics, 2011), and reconstructing the evolutionary dynamics of small RNAs (Genome Biology and Evolution, 2012). In an important review, Elizabeth defined genetic exchange communities (GECs), and considered what graphical structures might best represent them (FEMS Microbiological Reviews, 2011). In our Open Biology article, Elizabeth showed that for 27 E. coli and Shigella genomes, GECs were sculpted more by phyletic relatedness than by shared environment or lifestyle. Although we didn’t have the tools to take that further mechanistic step, her observation implicates recombination (into the genome) and integration (into control networks) more than, say, the availability of exogenous DNA per se.
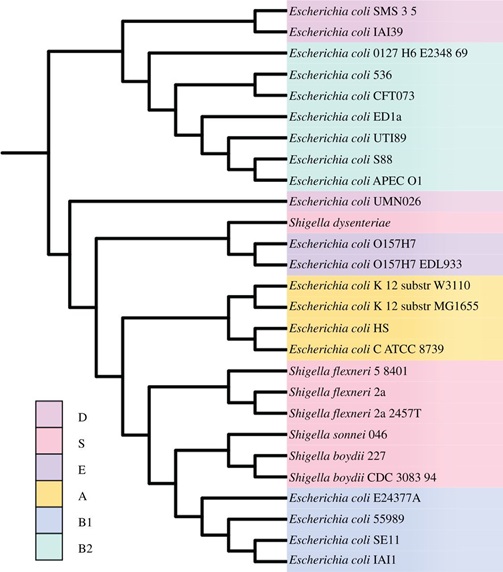
The title of our Open Biology article is precise, if a little peculiar. Construction isn’t something we, as computational biologists, were doing. GECs construct themselves, exploiting and modifying their real-world environments. As for bias, the Escherichia pan-genome is open: that is, it incorporates genes and gene fragments from well beyond just its close relatives. But in doing so, for these 27 strains at least, relatedness is more important than ecology or lifestyle.
[Elizabeth:] In 2012, I was wrapping up my thesis project and looking to the future. Shortly thereafter I headed to Bloomington, Indiana, where I had the opportunity to continue my education in gene transfer and genomics but this time focusing on plants rather than bacteria.
Mark: What has been a career highlight for you? How do you spend your free time after academic life?
Research isn’t the most carefree of careers, but I’ve found it hugely satisfying. Different colleagues think of me as a marine biologist, a computational biologist or bioinformatician, a biomedical scientist, a microbiologist or protistologist. It’s been a great privilege to study the natural world and its wondrous diversity of organisms. Mathematics and algorithms can be beautiful in their own right too, and computational approaches offer unique insights into real-world biology. A particular highlight has been to mentor, and learn from, talented students and postdocs – colleagues now, and our collective hope for the future.
After my alleged retirement at the end of 2017, I returned to a longstanding book-project on the history of the idea that there’s more to the living world than plants and animals. For over 2500 years, people have struggled to classify sponges, corals, zoophytes, Hydra, fungi, diatoms, protists, archaea... not to mention marvels such as the borametz. Watch for the book! (Oxford University Press, publication anticipated August 2022). And after that – I look forward to time with family and friends, travelling again, volunteer work with an environmental charity, and – as throughout – enjoying music.
Elizabeth, what career advice would you give to PhD students in a similar position to the one you were in ten years ago?
It is very difficult to give career advice because one of key the things I have taken away from the last ten years is that all careers are different and most advice won’t be right for everyone. I will say that openness to change, and a flexible mind, have served me well. They have afforded me the privilege of learning from inspiring mentors, and working on challenging, messy and beautifully surprising scientific questions.
As a graduate student in Australia, I could not have foreseen that ten years later I would be living and working in the Bay Area, routinely using technologies I could have only previously dreamed about. My happiest times in science have always been working with others, whether it has been sitting on the floor in a stack of papers with Mark trying to decide on the next question to tackle, or working with huge multi-disciplinary drug discovery teams. The advice I would give to my younger self is to always collaborate generously, travel whenever the opportunity presents itself, and prioritize physical and mental rest when you need to.
Open Biology celebrates ten years as the Royal Society’s open access journal for cellular and molecular biology. Find out more about becoming an author and how to submit.
Figure captions
'Mineral flowers' scanning electron microscopy (SEM) micrograph of minerals by Elena Tsolaki. 2019 Royal Society Publishing Photography Competition entry - Micro-imaging Category.
The E. coli–Shigella reference supertree. Open Bio. Doi: 10.1098/rsob.120112