Links to external sources may no longer work as intended. The content may not represent the latest thinking in this area or the Society’s current position on the topic.
Membrane pores: from structure and assembly, to medicine and technology
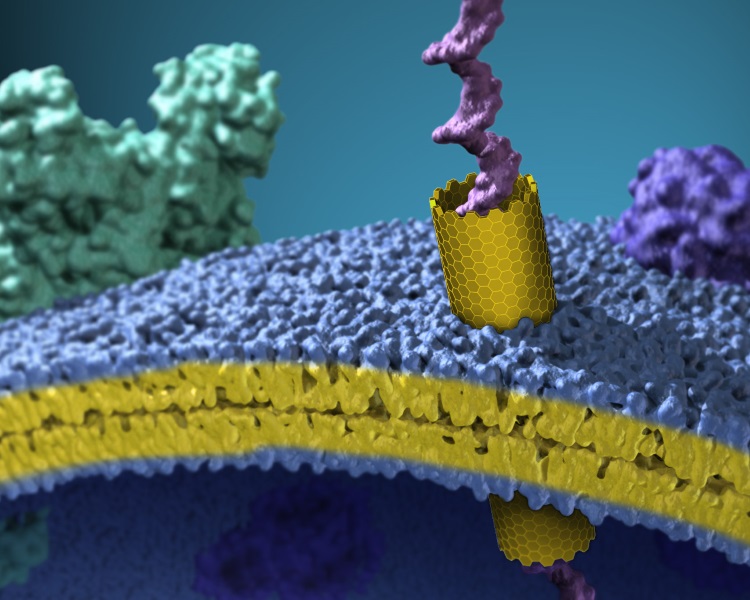
Scientific discussion meeting organised by Professor Robert Gilbert, Professor Hagan Bayley FRS and Professor Gregor Anderluh
Biological membranes define and protect living cells. Proteins can open aqueous pores through these barriers. Such pores play critical roles in infection, parasitism, immunity and neurodegeneration. This meeting will showcase our current understanding of the structure and function of membrane pores, the medical significance of pores, and how pore-forming proteins can be used in nanotechnological applications.
Speaker biographies, abstracts, and recorded audio of the presentations is available below. Papers from the meeting are available in an issue of Philosophical Transactions B.
Enquiries: Contact the events team.
Organisers
Schedule
Chair

Professor Gregor Anderluh, National Institute of Chemistry, Slovenia

Professor Gregor Anderluh, National Institute of Chemistry, Slovenia
Gregor Anderluh is director of the National Institute of Chemistry, Ljubljana, Slovenia. He received his PhD in Biology from the University of Ljubljana and did postdoctoral studies at the University of Newcastle, UK. He established an Infrastructural Centre for Molecular Interaction Analysis at the University of Ljubljana, Slovenia. He is studying protein-membrane interactions and how cellular membranes are damaged by proteins in bacterial pathogenesis and the immune system. His current interests are establishing novel biophysical approaches for studying protein binding to membranes and developing protein nanopores for applications.
| 09:00 - 09:30 |
Membrane pore-forming proteins in the molecular arms race between host and pathogen
Pathogens have evolved weapons to invade and damage our cells, and our immune system has evolved defences against these attacks. Among the weaponry used by both sides in this continual war are proteins that punch holes in cell membranes. Membrane perforation enables pathogens to take over host cells and resources for their own replication, and also enables host immune systems to kill invading pathogens. The membrane attack complex-perforin (MACPF)/ cholesterol dependent cytolysin (CDC) superfamily of membrane pore-forming proteins is used by a wide range of pathogens as well as by host immune systems. This talk focuses on the mechanisms by which MACPF and CDC proteins convert from their soluble, monomeric forms into large arcs and rings that insert into membranes and perforate them. Studies of pore assembly on liposomes in vitro and examples of their action in vivo will be presented. 
Professor Helen Saibil FMedSci FRS, Birkbeck College, UK

Professor Helen Saibil FMedSci FRS, Birkbeck College, UKHelen Saibil was educated in biophysics at McGill University, Montreal and then at Kings College London, where she did her PhD on the structure of retinal photoreceptor membranes. In her present position at Birkbeck College, her main interests are in analyzing conformational changes in macromolecular machines and the actions of intracellular pathogens on their hosts, using cryo-electron microscopy and image processing. These studies include work on protein folding, unfolding and disaggregation by molecular chaperones, and refolding during membrane pore formation by bacterial toxins and by pore forming proteins of the immune system. She is involved in setting up the national facility for cryo EM at the Diamond synchrotron, and is a member of EMBO, a Fellow of the Royal Society and a Fellow of the Academy of Medical Sciences. Web page: people.cryst.bbk.ac.uk/~ubcg16z/ |
|
|---|---|---|
| 09:30 - 10:00 |
Assembly mechanism of the pore-forming toxin Cytolysin A from Escherichia coli
The a-pore-forming toxin Cytolysin A (ClyA) is responsible for the hemolytic phenotype of several Escherichia coli and Salmonella enterica strains and is cytotoxic towards cultured mammalian cells. ClyA is a soluble, monomeric 34 kDa protein that spontaneously forms ring-shaped, homododecameric pore complexes upon contact with target membranes or detergent. The pore complex forms a hollow cylinder with an inner diameter of 70 Å that is reduced to 30 Å in the membrane-inserted part of the complex. The structural comparison between the soluble momoner and the protomer in the pore complex revealed one of the largest conformational transitions observed in a protein so far, in which more than half of the ClyA residues are reorganized and 16% of the residues are located in different secondary structure elements after protomer formation. Analysis of detergent-induced ClyA assembly in vitro showed that the rate-limiting step in pore formation at ClyA concentrations above 100 nM is the unimolecular transition of the soluble monomer to the assembly-competent protomer, which is followed by rapid protomer assembly. Single-molecule Förster resonance energy (FRET) experiments at picomolar ClyA concentrations preventing pore assembly showed that a molten globule-like off-pathway intermediate is formed during the monomer-to-protomer transition. The global kinetic analysis of detergent-induced pore formation from picomolar to micromolar ClyA concentrations supports a model in which all sterically compatible, linear oligomeric ClyA intermediates participate in pore assembly. 
Professor Rudolph Glockshuber, ETH Zurich, Switzerland

Professor Rudolph Glockshuber, ETH Zurich, SwitzerlandThe research of the Rudi Glockshuber focuses on mechanisms of protein folding and assembly. Besides general topics in protein folding such as catalysis of disulfide bond formation and slow folding reactions determined by proline cis-trans isomerization, the Glockshuber group investigates the assembly of supramolecular protein complexes, including the formation of amyloid beta fibrils and the assembly of virulence factors of bacterial pathogens such as adhesive type 1 pili and the pore poring toxin ClyA from Escherichia coli. |
|
| 10:00 - 10:25 | Discussion | |
| 10:25 - 10:50 | Coffee | |
| 10:50 - 11:20 |
Designing water-soluble and membrane-spanning peptide barrels
This talk will describe how we have developed an understanding of natural coiled-coil sequence motifs found in proteins, which lead to specific protein oligomerization; and then how we can apply this knowledge in rational peptide and protein design. For the latter, it is now clear that nature has almost certainly only shown us a glimpse of what is possible structurally and functionally with polypeptide chains. To help explore past the confines of natural coiled-coil structures, and into what has been termed the dark matter of protein space, a toolkit of de novo peptides has been developed. These can be used as building blocks for the rapid construction of new protein structures and assemblies. The talk will demonstrate the utility of this approach to make water-soluble protein-like barrels and pores, which can be engineered to bind biological molecules and catalyse simple reactions. More recently membrane-spanning variants of these α-helical barrels have been engineered, and in collaboration with the Bayley lab, it has shown that these insert into lipid bilayers and conduct ions in voltage-dependent manners. The two systems have been explored. In both cases, the insertion and orientation of peptides into the membranes can be directed, and the channel activities can be controlled through mutagenesis and blocking experiments. 
Professor Dek Woolfson, University of Bristol, UK

Professor Dek Woolfson, University of Bristol, UKProfessor Dek Woolfson took his first degree in Chemistry at the University of Oxford, UK. He then did a PhD at the University of Cambridge followed by post-doctoral research at University College London and the University of California, Berkeley. After 10 years as Lecturer through to Professor of Biochemistry at the University of Sussex, he moved to the University of Bristol in 2005 to take up a joint chair in Chemistry and Biochemistry. Dek’s research is at the interface between chemistry and biology, applying chemical methods and principles to understand biological phenomena. His group is interested in the challenge of rational protein design, and how this can be developed for synthetic biology. He places particular emphasis on making completely new protein structures and biomaterials for applications in cell biology and medicine. In 2011, Dek became the first recipient of the Medimmune Protein and Peptide Science Award of the Royal Society of Chemistry; and in 2014 he received a Royal Society Wolfson Research Merit Award. Dek is Director of BrisSynBio, a £13.6M BBSRC/EPSRC-funded Synthetic Biology Research Centre. |
|
| 11:20 - 11:50 |
Analysis of designed and natural proton transporters
The mechanism of proton transport through membrane proteins is of general interest to multiple areas of biology. Using a variety of spectroscopic, crystallographic, and computational methods, the mechanism by which protons are conducted through the M2 proton channel from influenza A virus was investigated, and this information was used to design new anti-influenza drugs that target highly drug-resistant forms of the virus. A second topic of the talk will focus on the use of de novo protein design to test the mechanism by which a class of transporters uses proton gradients to drive the conduction of molecules into or out of cells. Transporters have been hypothesized to arise by physical association or gene duplication of primordial units, leading to an assembly with “frustrated symmetry” that rocks between two states with the substrate-binding site alternately accessing each side of the membrane. Rocker, a minimalist Zn2+/proton antiporter was designed to test these principles, although it bears no sequence similarity to any known natural protein. Structural, dynamic and functional studies indicate that Rocker is a primordial transporter, which recapitulates many of the properties of this class of proteins. These studies also demonstrate the feasibility of using de novo design to design membrane proteins with complex dynamic and functional properties. 
Professor William DeGrado, University of California, San Francisco, USA

Professor William DeGrado, University of California, San Francisco, USAWilliam (Bill) DeGrado’s work focuses on the design of small molecule drugs, peptides, proteins, and peptide mimetics. Bill is currently a professor in the Department of Pharmaceutical Chemistry at the University of California San Francisco, where he is also a member of the Cardiovascular Research Institute. Before joining UCSF in 2011, he was a member of DuPont Central Research and DuPont Merck Pharmaceutical Company from 1981 to 1996 and the Raiziss Professor in the Department of Biochemistry and Biophysics at the University of Pennsylvania (1996 – 2011). Some of Bill’s research interests include: de novo design of proteins and peptides; peptide mimetics; structure/function of membrane proteins, including integrins and viral ion channels; small molecule drug design; bioinorganic chemistry. Bill is a member of the U.S. National Academy of Science, the U.S. National Academy of Inventors, the American Academy of Arts and Sciences, and a fellow of the American Association for the Advancement of Science. Bill graduated from Kalamazoo College in 1978, received his Ph.D. in organic chemistry from the University of Chicago (1981), and joined DuPont Central Research without an intervening postdoctoral position. |
|
| 11:50 - 12:15 | Discussion |
Chair

Professor Mauro Dalla Serra, National Research Council of Italy, Italy

Professor Mauro Dalla Serra, National Research Council of Italy, Italy
Mauro Dalla Serra is a Senior Research Scientist of the National Research Council of Italy and Head of the Unit at Trento of the Institute of Biophysics. His scientific research interest is to investigate the structural and functional aspects of the interaction of membrane-active molecules with natural and model lipid membranes. A large portion of his work has been dedicated to protein-protein and protein-lipid interaction, to understand the mechanism of action of pore-forming proteins, that are relevant for human health. He has published more than 90 peer reviewed articles and contributed 17 chapters to multi-authored books.
| 13:10 - 13:40 |
Structural basis for complement membrane attack complex formation
The membrane attack complex (MAC) is a fundamental component of immune defence that drills holes in bacterial membranes and kills pathogens. MAC lesions were first identified in 1964, yet half a century later details of its structure and assembly mechanism remain undiscovered. Here electron cryo-microscopy is used to visualize the human pore complex at subnanometer resolution. The protein composition of the MAC is determined and interaction interfaces that hold the assembly together are identified. Unlike closely related pore-forming proteins, the MAC’s asymmetric pore and "split-washer" shape suggest a killing mechanism that involves not only membrane rupture, but also distortion. 
Dr Doryen Bubeck, Imperial College London, UK

Dr Doryen Bubeck, Imperial College London, UKDoryen Bubeck received her PhD in Biophysics from Harvard University in 2005 where she used cryo-electron micrsocopy to investigate the cell entry mechanism of poliovirus. As an EMBO postdoctoral fellow and Cancer Research Institute Fellow at the University of Oxford, she continued to explore the structures of membrane proteins, focusing on the complement immune pathway. A recent highlight (Hadders & Bubeck et al., Cell Rep. 2012) is the discovery that complement components associate through a sideways alignment of their central MAC-perforin domains (MACPF). These results provide a structural framework for understanding the complex protein associations underlying activation of this innate immune effector. Doryen Bubeck is a lecturer in Structural Biology within the Department of Life Sciences and recipient of a Cancer Research UK Career Establishment Award. Her current research adopts an integrated structural approach merging cryo-electron microscopy and Xray crystallography to investigate the role of membrane proteins in host-pathogen interactions. She aims to investigate how membrane attack complex (MAC) pore formation is controlled, a process important for fighting infections and preventing complement-mediated tissue damage. |
|
|---|---|---|
| 13:40 - 14:10 |
Pore formation assisted by lipids
Pore-forming toxins (PFT) constitute a fascinating group of proteins belonging to the molecular offensive and defensive machinery of virtually all kingdoms of life. This class of water-soluble proteins shares the remarkable ability to metamorphose in the presence of cell membranes, generating lytic pores and causing cell-damage. Actinoporins are a family of potent hemolytic toxins from sea anemone forming alpha-helical pores on cellular and model membranes. In general, two requirements are sufficient to trigger pore-formation by actinoporins: (i) the presence of the lipid sphingomyelin, and (ii) the segregation of the membrane on domains or lipid-rafts. Until recently, the molecular basis of pore-formation by actinoporins, and specially the specific requirement for sphingomyelin were unclear. However, a number of recent studies have shed light into critical steps of their mechanism of action, such as binding of the toxins to the membrane, self-assembly via protein-protein interactions, and assembly of the transmembrane pore. Collectively, the data suggests that sphingomyelin facilitates pore-formation at the binding and assembly stages, and reveal the first example of a hybrid lipid/protein pore by a PFT. The structural and thermodynamic basis of this novel architecture will be explained in detail during this presentation. Surprisingly, the entire process can be made reversible under mild experimental conditions by the careful selection of detergents, challenging current perceptions in the field of membrane-protein interactions. 
Dr Jose Caaveiro, University of Tokyo, Japan

Dr Jose Caaveiro, University of Tokyo, JapanDr Caaveiro graduated in the University of the Basque Country (Spain), where he studied the modulation of peptides and proteins by lipids. After a postdoctoral period in USA (MIT and Brandeis University) he then joined the University of Tokyo (since 2008). Dr. Caaveiro’s research program combines structural and thermodynamic data to address a broad range of biologically inspired phenomena. His ultimate goal is to understand the physicochemical basis underpinning biomolecular recognition at the atomic scale. Recently, Dr. Caaveiro and co-workers has revealed an unprecedented role for lipids in the pore-forming mechanism of the potent hemolytic class of actinoporins. |
|
| 14:10 - 14:35 | Discussion | |
| 14:35 - 14:55 | Tea | |
| 14:55 - 15:25 |
New insights into Bax pore formation from advanced microscopy methods
Bax is a key player in apoptosis that mediates of the permeabilization of the outer mitochondrial membrane. Despite intense research, the underlying process remains poorly understood. By combining biophysical approaches at different scales, new insight into the molecular mechanism of Bax is provided. Electron paramagnetic resonance data shows a key conformational change in the central hairpin of Bax that is involved in pore formation. In this configuration, Bax is present as a mixture of oligomers based on dimer units, as revealed by single molecule imaging. Moreover, the nanoscale organization of Bax at mitochondria of apoptotic cells is provided by superresolution microscopy. Based on this, a new model for the molecular mechanism of Bax is proposed. 
Dr Ana-Jesus Garcia-Saez, University of Tübingen, Germany

Dr Ana-Jesus Garcia-Saez, University of Tübingen, GermanyAna-Jesus Garcia-Saez is academic background includes Professor (W3) at the Interfaculty Institute for Biochemistry (IFIB), Universität Tübingen, Germany (October 2013), Max Planck Research Group Leader and Deutsches Krebsforschungzentrum (DKFZ) Junior Group Leader. Bioquant, Germany (2010 - current), Post-doc at the BioTec, TU Dresden, Germany (2005-2009) and PhD at the Department of Biochemistry and Molecular Biology, University of Valencia, Spain (2000-2005) Fellowships and awards include: • European Research Council (ERC) Starting Grant (project acronym: APOQUANT), 2012-2017. |
|
| 15:25 - 15:55 |
Regulating Bak and Bax pore formation in the mitochondrial outer membrane
Two members of the Bcl-2 family, Bak and Bax, drive apoptotic cell death by changing conformation and forming oligomers that permeabilise the mitochondrial outer membrane. The two proteins are activated by BH3-only family members binding to the α2-α5 hydrophobic surface groove. Newly exposed hydrophobic regions then either “collapse” onto the membrane surface to lie in-plane, or interact to generate BH3:groove symmetric dimers. We found that in each Bak dimer the N-termini are fully solvent-exposed and mobile, allowing disulphide bonding between certain residues (e.g. V61C:V61C') to specifically interrogate how dimers associate into high order complexes. These data informed mathematical simulations that support a model in which Bak dimers interact in a random manner to form compact clusters that generate lipidic pores. It was also found that antibodies can trigger activation of Bak and mitochondrial Bax, and do so by binding to a new activation site, the α1-α2 loop. The mechanism of antibody-mediated Bak activation involves α1 dissociation, revealed by biochemical studies and a structural model of Fab bound to Bak. Intriguingly, antibodies to the α1-α2 loop in cytosolic Bax could block its translocation to mitochondria. These data thus identify the α1-α2 loop as a new target for regulating Bak and Bax apoptotic function. 
Dr Ruth Kluck, The Walter and Eliza Hall Institute of Medical Research, Australia

Dr Ruth Kluck, The Walter and Eliza Hall Institute of Medical Research, AustraliaDr Kluck is a cell biologist who has studied apoptotic cell death since her PhD at the Queensland Institute for Medical Research, in collaboration with John Kerr who discovered apoptosis. As a postdoc at the La Jolla Institute for Allergy and Immunology (1995-2002), she made the seminal discovery that Bcl-2 proteins inhibit apoptosis by blocking mitochondrial release of cytochrome c. In 2002, she moved to the Walter and Eliza Hall Institute for Medical Research in Australia where she has focused on the regulation and function of the Bak and Bax pore-forming proteins. |
|
| 15:55 - 16:20 | Discussion | |
| 16:20 - 16:50 |
Peptide-stabilized membrane pores: insights from simulations
The mechanism by which amphipathic peptides permeabilize biological membranes is not well understood. Do the peptides form well-defined pores or simply dissolve the membranes in a detergent-like fashion? What is the lifetime of these pores? How do the sequence and structure of these peptides determine their permeabilizing function? Both experiment and theory face formidable challenges in obtaining detailed information on such labile structures. We have used two theoretical approaches to study peptide-induced pore formation in lipid bilayers. The first treats water and lipids implicitly and the peptide in atomistic detail. This simplified representation allows one to obtain useful insights into protein-membrane interactions. Using this approach we showed that antimicrobial peptides bind more strongly to membrane pores than to the flat membrane, consistent with the idea that they stabilize them. The second approach is fully atomistic molecular dynamics simulations, some on the 10-microsecond timescale, starting from inserted peptide aggregates. Such simulations of melittin, magainin, PGLa, alamethicin, and protegrin have revealed interesting differences between these peptides and possible explanations of the observed synergy between some of them. 
Professor Themis Lazaridis, City College of New York, USA

Professor Themis Lazaridis, City College of New York, USAThemis Lazaridis received a Diploma in Chemical Engineering from Aristotle University in Thessaloniki, Greece in 1987. He went on to earn a Ph.D. in Chemical Engineering from the University of Delaware, USA. His Ph.D. research involved, among other topics, the development of a practical approach to calculate the entropy of hydration in water, elucidating the molecular origin of the unfavorable entropy of hydrophobic hydration. He was a postdoctoral fellow at Harvard University from 1992 to 1998, continuing to work on the statistical thermodynamics of solvation. He also worked on the energetics of protein folding and developed an implicit solvation model for proteins in solution. Since 1998 he has been a faculty member at the City College of New York. His current research interests focus on modeling protein-membrane interactions and especially the process of pore formation in membranes by antimicrobial peptides or protein toxins using implicit solvent models and all-atom simulations. |
|
| 16:50 - 17:00 | Discussion |
Chair

Professor Robert Gilbert, University of Oxford, UK

Professor Robert Gilbert, University of Oxford, UK
Robert Gilbert did his BSc in Molecular Biology and Biochemistry at Durham University and completed a PhD at the University of Leicester on pneumolysin, a pore-forming toxin from Streptococcus pneumoniae. He has continued to work with pneumolysin and related proteins, alongside developing interests in RNA biology and protein biophysics with particular relevance to virology, cancer and immunology. He is now Professor of Biophysics and Director of Graduate Studies in the Nuffield Department of Clinical Medicine at Oxford University, and Managing Editor of the European Biophysics Journal.
| 09:00 - 10:30 |
Punching giant holes in cells: The MACPF/CDC family of pore forming toxins
Membrane attack complex/perforin-like (MACPF) proteins are an excellent example of self-assembling, integral membrane proteins and comprise the largest superfamily of pore forming proteins. These pore forming proteins are an ancient family found in all kingdoms of life with diverse roles in immunity, venom toxicity, predator defense, pathogenesis and uncharacterized roles in development biology. Moreover the MACPF family share a common evolutionary ancestor with the CDC family of pore forming toxins from Gram positive bacteria. The over-arching mechanism of this MACPF/CDC superfamily starts with soluble monomeric proteins that recognise and assemble on the target membrane into a large ring-shaped oligomer. The oligomer can then undergo a massive conformational change to become an integral membrane protein complex. These membrane complexes range from 1 – 3 Mega Daltons in size and are capable of the passive transport of folded globular proteins. Recently, the field has started to tackle the outstanding questions of the MACPF field by combining single particle cryo-electron microscopy (SP cryo-EM), combined with cutting edge computational and biophysics techniques. The combined structural data of key distinct clades of the MACPF/CDC family now shed light on how the MACPF/CDC family evolved and help us understand the common mechanism of MACPF/CDC proteins. More importantly, by studying a diverse range of MACPF/CDC proteins, it is now possible to see how the common MACPF/CDC fold has evolved for specific activities within the organism and how the ancillary domains modulate the activity of the common MACPF/CDC fold. 
Dr Michelle Dunstone, Monash University, Australia

Dr Michelle Dunstone, Monash University, AustraliaDr Dunstone is currently an ARC Future Fellow and group leader in Monash University’s Department of Biochemistry & Molecular Biology. Dr Dunstone passion lies in discovering the conformational change that occurs in the Membrane Attack Complex / Perforin-like superfamily of pore forming proteins. Dr Dunstone was initially trained in crystallography in the laboratory of Prof Michael Parker at St. Vincent's Institute, Australia. Her original PhD project focused on the structure of C9 from the Membrane Attack Complex. Michelle continued researching the wider MAC/perforin family of pore forming proteins as a postdoctoral researcher at Monash University. In 2007 this her bioinformatic and crystallography research culminated in the first representative structure of a MAC/perforin-like protein. Her recent research on pleurotolysin explores the structure MACPF toxins by combining single particle cryo-electron microscopy, biophysics and bioinformatics. |
|
|---|---|---|
| 09:30 - 10:00 |
Mechanisms of cell-to-cell spread by Listeria monocytogenes
Listeria monocytogenes has a remarkable ability to disseminate within its host during infection. My laboratory is examining how these bacteria can spread from cell-to-cell using the actin-based motility mechanism powered by ActA. In a recent study we showed that ActA-mediated Arp2/3 recruitment promotes actin polymerization in the cytosol (comet tail formation) but is primarily not responsible for the formation of cell surface protrusions. Instead, we find that protrusion formation requires recruitment and activation of mDia formins. Thus, Listeria exploits two types of actin polymerization to enable cell-to-cell spread. We also showed that efferocytosis, the process by which dying/dead cells are removed by phagocytosis, promotes cell-to-cell spread by Listeria during infection. We showed that protrusion formation is associated with plasma membrane damage due to LLO’s pore-forming activity. LLO also promotes the release of bacteria-containing protrusions from the host cell, generating membrane-derived vesicles with exofacial PS. The PS-binding receptor TIM-4 contributes to efficient cell-to-cell spread by Listeria in macrophages in vitro and growth of these bacteria is impaired in TIM-4-/- mice. Thus, Listeria promotes its dissemination in a host by exploiting efferocytosis. 
Professor John Brumell, The Hospital for Sick Children, Canada

Professor John Brumell, The Hospital for Sick Children, CanadaDr Brumell completed undergraduate studies at the University of Western Ontario with a major in Biochemistry. He completed his PhD at the University of Toronto in 1997. His doctoral research focused on phosphoryation-dependent signaling mechanisms in human leukocytes that encounter bacteria. He extended these studies from 1997-1998 at Mt. Sinai Hospital in Toronto. His postdoctoral studies at the University of British Columbia from 1998-2002 examined how Salmonella subverts host cells to cause infection. In 2002 Brumell returned to Toronto where he established a laboratory at the Hospital for Sick Children. He is now a Senior Scientist in the Cell Biology program and a Co-Director of the SickKids Inflammatory Bowel Disease Centre. In 2014 Dr. Brumell was awarded the Pitblado Chair in Cell Biology. |
|
| 10:00 - 10:25 | Discussion | |
| 10:25 - 10:50 | Coffee | |
| 10:50 - 11:20 |
Pilus biogenesis at the outer membrane of bacterial pathogens
Gram-negative pathogens commonly exhibit adhesive pili on their surface that mediate specific attachment to the host. A major class of pili is assembled via the chaperone/usher (CU) pathway. Type 1 and P pili have served as model systems for the elucidation of the CU biosynthetic pathway. Pilus assembly requires a periplasmic chaperone (FimC and PapD for type 1 and P pili, respectively) and an outer-membrane assembly platform termed “usher” (FiimD and PapC for type 1 and P pili, respectively). CU pilus subunits are produced in the cytoplasm, translocated to the periplasm by the Sec translocation machinery, and then taken up by a chaperone to cross the periplasmic space to reach the outer-membrane. At the outer-membrane, chaperone-subunit complexes are recruited to an outer-membrane assembly platform, the usher, which orchestrates recruitment and polymerization of subunits. Previous work has elucidated the molecular basis of chaperone function. Recent progress has shed light into the mechanism of pilus subunit assembly at the usher, leading to the elucidation of the entire cycle of pilus subunit incorporation. 
Professor Gabriel Waksman FMedSci FRS, UCL and Birkbank College, UK

Professor Gabriel Waksman FMedSci FRS, UCL and Birkbank College, UKGabriel Waksman obtained his PhD in 1982 from the University of Paris. After a short spell in industry and a postdoctoral training at the Rockefeller University in New York, he joined the faculty of Washington University School of Medicine (St Louis, USA) in 1993. In 2000, he became the Alumni Endowed Professor of Biochemistry and Molecular Biophysics, and in 2002 was appointed the first Roy and Diana Vagelos Professor of Biochemistry and Molecular Biophysics. In 2003, he moved to London (UK) to take up the Joint Chair of Structural and Molecular Biology at University College London and Birkbeck College London. The same year, he was awarded a Wolfson-Royal Society Merit Award and was appointed the Head of the Institute of Structural and Molecular Biology at UCL/Birkbeck. In 2006, he was appointed to the Courtauld Chair in Biochemistry at UCL, became Head of the Department of Biochemistry and Molecular Biology (now Research Department of Structural and Molecular Biology) at UCL and was appointed Head of the School of Crystallography (now Department of Biological Sciences) at Birkbeck. He was elected to EMBO in 2007, a Fellow of the Academy of Medical Sciences in 2008, a Fellow of the Royal Society in 2012, and a member of the German Academy of Sciences in 2013, He maintains an active research programme in the Structural and Molecular Biology of Bacterial Secretion Systems funded by a senior investigator award from the Wellcome Trust, an Advanced ERC grant, and a programme grant from MRC. |
|
| 11:20 - 11:50 |
Structural insight into the biogenesis of beta barrel membrane proteins
β-barrel membrane proteins are essential for nutrient import, signaling, motility, and survival. In Gram-negative bacteria, the β-barrel assembly machinery (BAM) complex is responsible for the biogenesis of β -barrel membrane proteins, with homologous complexes found in mitochondria and chloroplasts. Structures of BamA, the central and essential component of the BAM complex, were determined from two species of bacteria: Neisseria gonorrhoeae and Haemophilus ducreyi. BamA consists of a large periplasmic domain attached to a 16-strand transmembrane β-barrel domain. Three structural features speak to the mechanism by which BamA catalyzes β-barrel assembly. The first is that the interior cavity is accessible in one BamA structure and conformationally closed in the other. Second, an exterior rim of the β-barrel has a distinctly narrowed hydrophobic surface, locally destabilizing the outer membrane. And third, the β-barrel can undergo lateral opening, evocatively suggesting a route from the interior cavity in BamA into the outer membrane. A new structure of the BamACDE complex illustrates how the BamC, BamD, and BamE lipoproteins assemble on the BamA periplasmic domain and provides further evidence for lateral opening of the β -barrel. 
Dr Susan Buchanan, National Institute of Health, USA

Dr Susan Buchanan, National Institute of Health, USADr Buchanan is Chief of the Section on Structural Biology of Membrane Proteins in the National Institute for Diabetes & Digestive & Kidney Diseases, at the National Institutes of Health. She received her Ph.D. from the Johann-Wolfgang-Goethe Universität in Frankfurt, Germany in 1990. She completed postdoctoral fellowships at the MRC Laboratory of Molecular Biology, Cambridge, UK, and at UT Southwestern Medical Center, Dallas, before returning to the UK to establish a research group at Birkbeck College, London in 1998. She joined the NIDDK as a tenure track investigator in 2001 and is currently a Senior Investigator and Chief of the Laboratory of Molecular Biology, NIDDK. Research in Dr Buchanan’s laboratory focuses on the structure determination of integral membrane proteins by X-ray crystallography and functional analysis of these proteins using biophysical, biochemical, and cell biological techniques. Current topics include small molecule and protein import across the bacterial outer membrane, protein secretion by pathogenic bacteria, and protein import across mitochondrial outer membranes. |
|
| 11:50 - 12:15 | Discussion |
Chair

Professor Hagan Bayley FRS, University of Oxford, UK

Professor Hagan Bayley FRS, University of Oxford, UK
Hagan Bayley is the Professor of Chemical Biology at the University of Oxford. Major interests of his laboratory are the development of engineered pores for stochastic sensing, the study of covalent chemistry at the single molecule level, ultrarapid DNA sequencing and the fabrication of synthetic tissues. In 2005, Professor Bayley founded Oxford Nanopore to exploit the potential of stochastic sensing technology. The company has developed the MinION portable DNA sequencer. His research and entrepreneurial skills have been recognised several times. Professor Bayley was the 2009 Chemistry World Entrepreneur of the Year. In 2011, he was elected a Fellow of the Royal Society. In 2012 he was awarded the Royal Society of Chemistry's Interdisciplinary Prize, in 2017 the Menelaus Medal of the Learned Society of Wales and in 2019 the Mullard Award of the Royal Society. In 2018, Professor Bayley held the Kavli Chair at the Delft University of Technology.
http://bayley.chem.ox.ac.uk/
| 13:10 - 14:40 |
From DNA-based pores toward membrane motors
It is notoriously difficult to observe, let alone control, the position and orientation of molecules because of their small size and the constant thermal fluctuations that they experience in solution. Molecular self-assembly with DNA provides a route for placing molecules and constraining their fluctuations in user-defined ways and with up to Angstroem-scale precision. These positioning options open attractive and unprecedented avenues for scientific and technological exploration, in particular with respect to the creation of artificial membrane pores membrane-embedded mechanisms. This talk will introduce some of the key concepts and methods, and highlight a number of recent developments. 
Professor Hendrik Dietz, Technische Universität München, Germany

Professor Hendrik Dietz, Technische Universität München, GermanySince 2009 Hendrik Dietz has been a Professor for Biophysics and Principal Investigator of the Laboratory for Biomolecular Nanotechnology at Technische Universität München. Previously he was a Postdoctoral research Fellow at the Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School. Prizes include Gottfried Wilhelm Leibniz-Preis, Deutsche Forschungsgemeinschaft (2015), Engelhorn Forschungspreis, Peter-und-Traudl-Engelhorn Stiftung (2012), Hoechst Dozentenstipendium, Aventis Foundation and Fonds der Chemischen Industrie (2012), Arnold Sommerfeld Award, Bayerische Akademie der Wissenschaften (2010) and European Research Council Starting Grant (2010). |
|
|---|---|---|
| 13:40 - 14:00 |
Carbon Nanotube Porins: A biomimetic membrane pore for nanofluidics studies
Living systems control transport of ions or small molecules across biological membranes using ion channels that form pores in lipid bilayers. Although bottom-up synthesis and top-down fabrication could produce pores of comparable size, an unresolved challenge remains to build a simplified membrane nanopore scaffold. Carbon nanotube porins (CNTPs) formed by ultra-short carbon nanotubes (CNTs) modified with lipid surfactants show a set of behaviors that come remarkably close to imitating biological nanopores. The defining features of these nanostructures are their inner pores that have atomically smooth hydrophobic walls, which confine water on a molecular level, and, in some cases, down to a single-file configuration. We present experimental results that use bulk-scale and individual nanopore measurements to explore the physical origins of efficient transport in CNT porins, and focus on the role that molecular confinement plays in determining the transport characteristics and selectivity of the pore for water, protons, and ions. Overall, CNT porins represent a simplified biomimetic system that is ideal for studying fundamentals of nanofluidic transport and transport in biological channels, and for building complex engineered mesoscale structures. 
Dr Alex Noy, Lawrence Livermore National Laboratory, USA

Dr Alex Noy, Lawrence Livermore National Laboratory, USAAlex Noy is a Senior Research Scientist at LLNL. He joined LLNL in 1998 as the Lab’s first E.O. Lawrence Fellow after getting his Ph.D. from Harvard University. His current research interests center on biophysics, biomaterials, and nanoscience with a common theme of using one-dimensional nanomaterials to build biomimetic and bioengineered structures. His research group is currently active in the areas of (1) bioelectronics, where they develop devices that combine nanomaterials with biological and biomimetic components to with a goal to create a seamless bidirectional neural interface, (2) nanofluidics and biomimetic molecular transport, where they develop carbon nanotube porins: artificial membrane channels based on carbon nanotubes, and (3) high speed atomic force microscopy, where they aim to image biological processes in-situ in real time. Noy has also been an Adjunct Associate Professor at the School of Natural Sciences at the University of California Merced since 2005. |
|
| 14:00 - 14:25 | Discussion | |
| 14:25 - 14:55 | Tea | |
| 14:55 - 15:25 |
Toroidal pore formation in lipid membranes
A wide range of important biological processes involve the formation of toroidal pores in lipid membranes. Here we employ a combination of single-molecule fluorescence imaging and optical Single Channel Recording in Droplet Interface Bilayers to help understand the biophysics of this process. We image the formation of individual punctate mobile defects both in pure lipid bilayers, and in bilayers where this process if mediated by the presence of a range of peptides and proteins. 
Professor Mark Wallace, King's College London, UK

Professor Mark Wallace, King's College London, UKMark studied Chemical Physics as an undergraduate at the University of Bristol, followed by a PhD in Chemistry at the University of Cambridge under the supervision of Professor David Klenerman. He was awarded the 2002 Gregorio Weber International Prize in Biological Fluorescence for this work. Mark then spent 2 years as a postdoctoral fellow at Stanford University, working with Professor Richard Zare. He returned to the UK in 2002 to undertake a second postdoctoral position at the National Institute for Medical Research with Dr Justin Molloy, before moving to Oxford in 2005 as a Royal Society University Research Fellow. He was subsequently appointed as a University lecturer and fellow of Wadham College in 2006. He joined King’s in 2016 as part of its expansion of Chemistry. In 2009 Mark was appointed to the steering committee of the British Biophysical Society. Patents arising from his work have also been licensed in the UK Mark has also been active in raising awareness of his group’s research beyond the lab, including video podcasting and participation in the 2014 “I’m a scientist get me out of here” competition. He was awarded the RSC Norman Heatley Award in 2015 in recognition of his work. He was made a Fellow of the RSC in the same year. |
|
| 15:25 - 15:55 |
Single-protein analysis with a nanopore test-tube
The specific modulation of the ionic current through biological nanopores reconstituted in artificial membranes has been used to identify and analyze individual molecules. Here we show that ClyA, a biological nanopores with a 6.5 nm diameter and a 13 nm length, can be used as a nanoscale test-tube to study proteins. We found that the electrophoretic forces inside the nanopore can be used to trap and orient individual proteins for tens of minutes. The binding of analytes to the internalized protein is then reported by specific ionic current blockades. Remarkably, despite the diffusion of charged analytes inside the nanopore is strongly affected by the electric field, the binding constants for proteins inside the nanopore are identical to the values measured in bulk, indicating that the confinement and the electrophoretic forces inside the nanopore do not alter the folding and the binding properties of proteins. This technique will allow the label-free investigation of enzymatic reaction at the single molecule level, while nanopores with internal protein sensors might be incorporated into low-cost and portable analytical devices. 
Dr Giovanni Maglia, University of Groningen, Netherlands

Dr Giovanni Maglia, University of Groningen, NetherlandsGiovanni Maglia was born in Bologna (Italy). He attended the University of Bologna for his undergraduate studies in Pharmaceutical Chemistry, and went to the University of Pennsylvania (USA) for his Masters thesis. He obtained his PhD from the University of Birmingham (UK) investigating the physical bases of the hydride transfer reaction catalyzed by dihydrofolate reductase. After a short spell at the university of Leuven (BE), he moved to the University of Oxford for his post-doctoral research on DNA nanopore sequencing. In 2010 Giovanni Maglia received an ERC starting grant to initiate his independent research at the University of Leuven (BE). From November 2011, he is associate professor of Chemical Biology at the University of Groningen (NL). His research interests include single-molecule enzymology, single-molecule proteomics and sensing, and designing and engineering artificial transmembrane machines. |
|
| 15:55 - 16:20 | Discussion | |
| 16:20 - 16:50 |
Mass spectrometry of membrane pores - the lipid connection
The realization that the lipid environment is crucial for maintaining the structure and function of membrane proteins prompts new methods to understand lipid interactions. One such method, mass spectrometry, is emerging with the potential to monitor different modes of lipid binding to membrane protein complexes. Initial studies monitored the addition of lipids and deduced the kinetic and thermodynamic effects of lipid binding to proteins. Recent studies have focused on identifying lipids already present, explicitly in plugs, annular rings or cavities. Lipids that bind within these orifices to membrane proteins will have higher residence times than those in the bulk lipid bilayer and consequently can be quantified and characterized by mass spectrometry. In special cases, lipids identified within cavities have been proposed as substrates following activity assays. Alternatively, a gas phase unfolding protocol can be used to distinguish lipids that are important for stability. This lecture will provide an overview of recent advances in mass spectrometry, with a particular focus on the distinction of the various modes of lipid binding and their implications for structure and function of membrane pores. 
Dame Carol Robinson FMedSci FRS, University of Oxford, UK

Dame Carol Robinson FMedSci FRS, University of Oxford, UKCarol Robinson has pioneered the use of mass spectrometry for structural biology and biophysics, with applications to a broad spectrum of biologically important macromolecular assemblies. Recent highlights from her work include the discovery that membrane protein complexes can be liberated from micelles in the gas phase while retaining their subunits interactions, lipid binding properties and overall topology. Carol currently holds the Chair of Doctor Lee’s Professor of Chemistry at the University of Oxford. Her research has attracted international awards and prizes including the Anfinsen Award from the Protein Society, the Biemann Medal from the American Society of Mass Spectrometry, the Davy Medal and the Rosalind Franklin Award from the Royal Society, the HUPO Award for Distinguished Achievement in Proteomic Science, and the Anatrace Award for Membrane Proteins from the American Biophysical Society. Carol also holds three honorary doctorates and received a DBE in 2013 for her contribution to science. |
|
| 16:50 - 17:00 | Discussion |
