Links to external sources may no longer work as intended. The content may not represent the latest thinking in this area or the Society’s current position on the topic.
SIMposium: recent advancements in structured illumination microscopy
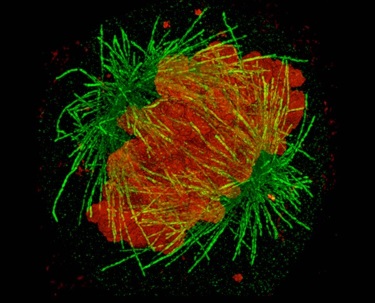
Theo Murphy scientific meeting organised by Dr Kirti Prakash, Dr Benedict Diederich, Professor Lothar Schermelleh, Dr Stefanie Reichelt and Professor Rainer Heintzmann.
Structured illumination microscopy (SIM) has emerged as an essential super-resolution technique for 3D and live-cell imaging. However, to date, there has not been a dedicated symposium/workshop covering different aspects of SIM, from biological applications, use of commercial instruments, to bespoke hardware and software development. The meeting aimed to recap recent developments as well as outline future trends.
A two-part Philosophical Transactions A journal issue accompanies this meeting. The first volume can be read here and the second volume can be read here.
Watch the event
Click watch on YouTube to view the full video playlist.
Enquiries: contact the Scientific Programmes team.
Organisers
Schedule
Chair

Dr Kirti Prakash, The Institute of Cancer Research, UK.

Dr Kirti Prakash, The Institute of Cancer Research, UK.
Kirti Prakash is a computer scientist by training (Bachelors and Masters degree) but a biologist at heart (PhD degree). Kirti aspires to be an inventor and develop new imaging tools for cell biology and neuroscience. Kirti did his Masters in Computer Science from Aalto University (Finland) and PhD in Biology from Heidelberg University (Germany). During his PhD, he developed a new method to image DNA which led to the first high-resolution images of the epigenetic landscape of meiotic chromosomes and mechanisms behind chromosome condensation. The doctoral research earned him several awards including Springer Best PhD Thesis Prize. After his PhD, he did a couple of postdocs at Carnegie Institution for Science (USA) and University of Cambridge (UK). The primary highlights of his research here were laser-free superresolution microscopy and the development of a high-content imaging pipeline to quantify single-cell gene expression. He then spent a couple of years at the National Physical Laboratory (UK) to work on standardisation and quality control of quantitative microscopy. Presently at the Institute of Cancer Research (UK), he is working on artificial intelligence and digital pathology.
| 08:45 - 09:00 | Welcome by the Royal Society & lead organiser |
|---|---|
| 09:00 - 09:30 |
Keynote: Thoughts on structured illumination - past, presence and future
This talk presents answers to fundamental questions related to structured illumination (SIM) and more generally superresolution microscopy, based on the speaker personal views on superresolution in light microscopy. He will discuss the definition of superresolution, Abbe's resolution limit and the classification of superresolution methods into nonlinear-, prior knowledge- and near-field-based superresolution. A further focus is put on the capabilities and technical aspects of present and future SIM methods. 
Professor Rainer Heintzmann, Institute of Physical Chemistry, Friedrich-Schiller-Universität Jena, Germany

Professor Rainer Heintzmann, Institute of Physical Chemistry, Friedrich-Schiller-Universität Jena, GermanyProfessor Rainer Heintzmann studied at the Universities of Osnabrück and Heidelberg and worked as a postdoctoral fellow at the Max Planck Institute of biophysical Chemistry in Göttingen and as a group at the Randall Division, King’s College London. He is currently professor of physical chemistry at the Friedrich-Schiller University Jena, and heads the microscopy research unit at the Leibniz Institute of Photonic Technology in Jena, Germany. His research focuses on methods for imaging cellular function at high resolution and developing techniques to measure multidimensional information in small biological objects such as cells, cellular organelles or other small structures of interest. Examples of his developments are structured illumination, image inversion interferometry, optical photon reassignment, and pointillism. He is highly interested is in computer-based reconstruction and inverse modelling methods such as deconvolution. |
| 09:30 - 10:00 |
SIM: a counterfactual history
SIM has undergone much improvement over the past 20 years, transforming from a technique capable of 2D imaging of fixed samples into ones capable of 3D live cell imaging, isotropic 100-nm resolution, and lateral resolutions finer than 65 nm. But what might have happened if the first SIM developments had never occurred? In this talk, Dr Manton considers how SIM might have arisen in a parallel universe and how we could have arrived at the current state-of-the-art through different approaches. The talk will look at developing both 2D and 3D SIM and then show how these approaches can be further developed to solve some outstanding problems in our universe. 
Dr James Manton, MRC Laboratory of Molecular Biology, Cambridge, UK

Dr James Manton, MRC Laboratory of Molecular Biology, Cambridge, UKJames is a physicist at the MRC Laboratory of Molecular Biology, where he works on developing new optical microscopy techniques for the study of biological samples. He has a particular interest in structured illumination and light sheet fluorescence microscopy, with a view towards rapid live imaging. |
| 10:00 - 10:30 |
Studying chromatin organisation and RNA dynamics by 3D-SIM
Super-resolution microscopy has undergone a remarkable evolution over the past two decades. However, despite its promise, it has not always delivered when it comes to widespread application in biology labs. In his talk Dr Schermelleh will present some of his and his team's efforts to make interference-based 3D structured illumination microscopy (3D-SIM) more reproducibly applicable to address open biological questions and use it as a genuine tool for new discoveries. Dr Schermelleh will showcase his and his team's recent work on analysing functional chromatin topography on the size scale of TADs, and studying Xist RNA dynamics during X chromosome inactivation in vivo and in situ, highlighting some of the technical advances in fluorescence labelling, and advanced image analysis that were required along the way. 
Dr Lothar Schermelleh, University of Oxford, UK

Dr Lothar Schermelleh, University of Oxford, UKLothar Schermelleh has studied Biology at the Ludwig Maximilian University Munich (LMU). He obtained his doctorate under the supervision of Thomas Cremer in 2003 for his work on the 'Dynamic organisation of chromosomes in the mammalian cell nucleus'. He later joined the group of Heinrich Leonhardt as a Postdoctoral Researcher and Lecturer, where he studied the role and regulation of DNA methylation 1 through developing and applying advanced single-cell imaging and analyses approaches. During this time, as a visiting scientist in the lab of John Sedat at the UCSF, he first came in contact with super-resolution structured illumination microscopy (SIM), and he later established this technique and its cell biological application at the LMU Munich. In 2011, Lothar joined the University of Oxford as Micron Senior Research Fellow and Principal Investigator at the Department of Biochemistry, where his group further advanced the development of quantitative super-resolution 3D imaging to study mesoscale chromatin organisation and mechanisms of epigenetic gene regulation, eg in X chromosome inactivation. In 2020, he was named Associate Professor and became Academic Director of the Micron Bioimaging Facility. |
| 10:30 - 11:00 | Coffee |
| 11:00 - 11:30 |
Structured illumination microscopy and image scanning microscopy: A comparison
Image formation in structured illumination microscopy (SIM) and image scanning microscopy (ISM) is compared. Image scanning microscopy is a confocal microscopy technique using a detector array in place of the usual confocal pinhole with a single element detector. Both techniques can result in a doubling of spatial frequency bandwidth compared with conventional fluorescence microscopy. The performance of SIM is usually presented in a way that includes digital reconstruction, which makes comparison with other techniques difficult. Here, the researchers consider the resolution of SIM based on the optical properties alone, and compare it with ISM using pixel reassignment. SIM results in a superior resolution. Of course, in both cases digital processing can result in further improvements in resolution. The main advantage of ISM over SIM is the optical sectioning property, which allows ISM to penetrate better through thick specimens. 
Dr Colin Sheppard, University of Wollongong and Italian Institute of Technology, Australia and Italy

Dr Colin Sheppard, University of Wollongong and Italian Institute of Technology, Australia and ItalyColin Sheppard obtained his PhD in Engineering from University of Cambridge (1973), and DSc degree in Physical Sciences from University of Oxford (1986). Professor Sheppard is currently Honorary Professorial Fellow at University of Wollongong, Australia; External Collaborator and Visiting Scientist with Italian Institute of Technology, Genoa; and Supernumerary Fellow of Pembroke College Oxford. He was Lecturer in Engineering at University of Oxford, and Fellow of Pembroke College; Professor of Physics and Head of Physical Optics Department at University of Sydney, Australia; Professor and Head of Department of Bioengineering at the National University of Singapore; and Senior Scientist at Italian Institute of Technology in Genoa. He was elected Fellow of OSA, SPIE, IEE, Institute of Physics, Japan Society for Applied Physics, and Royal Microscopical Society. He was Vice-President of International Commission for Optics, and President of International Society for Optics Within Life Sciences (OWLS). He received Italian Society for Optics and Photonics Galileo Galilei Medal, Institute of Physics Optics and Photonics Division Prize, Max Planck Bonhöffer Medal, Humboldt Research Award, ISATA Mercedes Award, National Physical Laboratory Metrology Award, Prince of Wales Award for Industrial Innovation, British Technology Group Academic Enterprise Award, and IEE Gyr and Landis Prize. |
| 11:30 - 12:00 |
Overcoming physical resolution limits of fluorescence microscopes with sparse deconvolution
To enable live-cell long-term super-resolution (SR) imaging, Chen’s group have developed a deconvolution algorithm for structured illumination microscopy based on Hessian matrixes (Hessian-SIM). It uses the continuity of biological structures in multiple dimensions as a priori knowledge to guide image reconstruction and attains artifact-minimised SR images with less than 10% of the photon dose used by conventional SIM while substantially outperforming current algorithms at low signal intensities. Its high sensitivity allows the use of sub-millisecond excitation pulses followed by dark recovery times to reduce photobleaching of fluorescent proteins, enabling hour-long time-lapse SR imaging in live cells. Thereafter, they take advantage of a priori knowledge of the sparsity and continuity of fluorescently labeled biological structures, and develop a deconvolution algorithm that further extends the resolution of super-resolution microscopes under the same photon budgets by nearly twofold. As a result, sparse structured illumination microscopy (Sparse-SIM) achieves ~60 nm resolution at a 564 Hz frame rate, allowing it to resolve intricate structural intermediates, including small vesicular fusion pores, ring-shaped nuclear pores formed by different nucleoporins, and relative movements between the inner and outer membranes of mitochondria in live cells. Likewise, sparse deconvolution can be used to increase the three-dimensional resolution and contrast of spinning-disc confocal-based SIM (SD-SIM), and operates under conditions with the insufficient signal-to-noise ratio, all of which allows routine four-color, three-dimensional, ~90 nm resolution live-cell super-resolution imaging. Overall, sparse deconvolution may be a general tool to push the spatiotemporal resolution limits of live-cell fluorescence microscopy. 
Dr Liangyi Chen, Peking University, China

Dr Liangyi Chen, Peking University, ChinaLiangyi Chen is Boya Professor of Peking University. He obtained his undergraduate degrees Biomedical engineering in Xi’an JiaoTong University, then majored in Biomedical engineering in pursuing PhD degree in Huazhong University of Science and Technology. His lab focused on two interweaved aspects: the development of new imaging and quantitative image analysis algorithms, and the application of these technology to study how glucose-stimulated insulin secretion is regulated in the health and disease at multiple levels (single cells, islets and in vivo) in the health and disease animal models. The techniques developed included ultrasensitive Hessian structured illumination microscopy (Hessian SIM) for live cell super-resolution imaging, the Sparse deconvolution algorithm for extending spatial resolution of fluorescence microscopes limited by the optics, Super-resolution fluorescence-assisted diffraction computational tomography (SR-FACT) for revealing the three-dimensional landscape of the cellular organelle interactome, two-photon three-axis digital scanned lightsheet microscopy (2P3A-DSLM) for tissue and small organism imaging, and fast High-resolution Miniature Two-photon Microscopy (FHIRM-TPM) for Brain Imaging in Freely-behaving Mice. He is also recipient of the National Distinguish Scholar Fund project from National Natural Science Foundation of China. |
| 12:00 - 12:45 |
Panel discussion: Open challenges in SIM data acquisition and processing

Professor Rainer Heintzmann, Institute of Physical Chemistry, Friedrich-Schiller-Universität Jena, Germany

Professor Rainer Heintzmann, Institute of Physical Chemistry, Friedrich-Schiller-Universität Jena, GermanyProfessor Rainer Heintzmann studied at the Universities of Osnabrück and Heidelberg and worked as a postdoctoral fellow at the Max Planck Institute of biophysical Chemistry in Göttingen and as a group at the Randall Division, King’s College London. He is currently professor of physical chemistry at the Friedrich-Schiller University Jena, and heads the microscopy research unit at the Leibniz Institute of Photonic Technology in Jena, Germany. His research focuses on methods for imaging cellular function at high resolution and developing techniques to measure multidimensional information in small biological objects such as cells, cellular organelles or other small structures of interest. Examples of his developments are structured illumination, image inversion interferometry, optical photon reassignment, and pointillism. He is highly interested is in computer-based reconstruction and inverse modelling methods such as deconvolution. 
Dr James Manton, MRC Laboratory of Molecular Biology, Cambridge, UK

Dr James Manton, MRC Laboratory of Molecular Biology, Cambridge, UKJames is a physicist at the MRC Laboratory of Molecular Biology, where he works on developing new optical microscopy techniques for the study of biological samples. He has a particular interest in structured illumination and light sheet fluorescence microscopy, with a view towards rapid live imaging. 
Dr Marcel Müller, Bielefeld University, Germany

Dr Marcel Müller, Bielefeld University, GermanyMarcel Müller works on implementing microscope systems and data reconstruction algorithms for structured illumination microscopy (SIM). In 2016, he started the fairSIM project, which aims to provide SIM-related resources (software plugins, example datasets, system blueprints) to the scientific community. He has since worked at the Micron Advanced Imaging Facility at the University of Oxford, as a MSCA fellow at the KU Leuven, and is currently a Postdoctoral Researcher at Bielefeld University in Germany. His research focused on new, robust, fast and cost-effective implementations of structured illumination microscopy. 
Dr Reto Fiolka, The University of Texas Southwestern Medical Center, USA

Dr Reto Fiolka, The University of Texas Southwestern Medical Center, USATrained as a mechanical engineer in computational fluid dynamics, Reto Fiolka completed his PhD at the institute of Nanotechnology at ETH Zurich in the group of Dr Andreas Stemmer. He conducted post-doctoral research at the Howard Hughes Medical Institute's Janelia Research Campus under the late Dr Mats Gustafsson working on 3D structured illumination microscopy and under Dr Meng Cui on adaptive optics. At UT Southwestern, the research in his lab aims to extend the current imaging capabilities of optical microscopy such that cancer cell research and drug screening can be performed in physiologically relevant, 3D environments, ex vivo and in vivo. His microscope development is focused on improving the spatiotemporal resolution and optical penetration depth and translating new technologies to biological research. 
Dr Colin Sheppard, University of Wollongong and Italian Institute of Technology, Australia and Italy

Dr Colin Sheppard, University of Wollongong and Italian Institute of Technology, Australia and ItalyColin Sheppard obtained his PhD in Engineering from University of Cambridge (1973), and DSc degree in Physical Sciences from University of Oxford (1986). Professor Sheppard is currently Honorary Professorial Fellow at University of Wollongong, Australia; External Collaborator and Visiting Scientist with Italian Institute of Technology, Genoa; and Supernumerary Fellow of Pembroke College Oxford. He was Lecturer in Engineering at University of Oxford, and Fellow of Pembroke College; Professor of Physics and Head of Physical Optics Department at University of Sydney, Australia; Professor and Head of Department of Bioengineering at the National University of Singapore; and Senior Scientist at Italian Institute of Technology in Genoa. He was elected Fellow of OSA, SPIE, IEE, Institute of Physics, Japan Society for Applied Physics, and Royal Microscopical Society. He was Vice-President of International Commission for Optics, and President of International Society for Optics Within Life Sciences (OWLS). He received Italian Society for Optics and Photonics Galileo Galilei Medal, Institute of Physics Optics and Photonics Division Prize, Max Planck Bonhöffer Medal, Humboldt Research Award, ISATA Mercedes Award, National Physical Laboratory Metrology Award, Prince of Wales Award for Industrial Innovation, British Technology Group Academic Enterprise Award, and IEE Gyr and Landis Prize. 
Dr Sara Abrahamsson, University of California Santa Cruz, USA

Dr Sara Abrahamsson, University of California Santa Cruz, USASara Abrahamsson is an Assistant Professor at UCSC pursuing groundbreaking research in optical systems design and diffractive optics for 3D microscopy and super-resolution microscopy. She has developed the imaging methods aberration-corrected Multifocus Microscopy and MultiFocus Structured Illumination Microscopy which enable live simultaneous 3D imaging at and beyond the diffraction limit of resolution. |
Chair

Dr Lothar Schermelleh, University of Oxford, UK

Dr Lothar Schermelleh, University of Oxford, UK
Lothar Schermelleh has studied Biology at the Ludwig Maximilian University Munich (LMU). He obtained his doctorate under the supervision of Thomas Cremer in 2003 for his work on the 'Dynamic organisation of chromosomes in the mammalian cell nucleus'. He later joined the group of Heinrich Leonhardt as a Postdoctoral Researcher and Lecturer, where he studied the role and regulation of DNA methylation 1 through developing and applying advanced single-cell imaging and analyses approaches. During this time, as a visiting scientist in the lab of John Sedat at the UCSF, he first came in contact with super-resolution structured illumination microscopy (SIM), and he later established this technique and its cell biological application at the LMU Munich. In 2011, Lothar joined the University of Oxford as Micron Senior Research Fellow and Principal Investigator at the Department of Biochemistry, where his group further advanced the development of quantitative super-resolution 3D imaging to study mesoscale chromatin organisation and mechanisms of epigenetic gene regulation, eg in X chromosome inactivation. In 2020, he was named Associate Professor and became Academic Director of the Micron Bioimaging Facility.
| 13:30 - 14:00 |
Keynote: Light-sheet microscopy with multi-directional structured illumination
Structured illumination microscopy (SIM) doubles the spatial resolution of a microscope without requiring high laser power or specialised fluorophores. In its 3D form, SIM illuminates the entire sample, which can lead to increased photo-bleaching and reconstruction artifacts due to out-of-focus blur. In contrast, light-sheet fluorescence microscopy (LSFM) mostly avoids exciting out-of-focus fluorescence and thereby drastically lowers photo-bleaching. While LSFM excels at long-term or high-speed volumetric imaging, its spatial resolution is modest. Therefore, combining LSFM with SIM appears attractive, as it promises gentle live cell imaging at doubled resolution. Unfortunately, combining the two is rather complex, as SIM requires illuminating the sample with three different pattern orientations. Applied to LSFM, this would require up to three illumination objectives to deliver differently oriented structured light-sheets. Here the researchers implement SIM in oblique plane microscopy (OPM), a light-sheet technique that requires only one objective for illumination and fluorescence detection. Rotation of the structured light-sheet is facilitated via a high-speed image rotator, which also de-rotates the fluorescence light for subsequent detection. This allows the researchers to double the resolution of OPM and image at up to 2Hz volumetric rate. They present imaging of the cytoskeleton, mitochondria dynamics and clathrin mediated endocytosis using this new microscope. 
Dr Reto Fiolka, The University of Texas Southwestern Medical Center, USA

Dr Reto Fiolka, The University of Texas Southwestern Medical Center, USATrained as a mechanical engineer in computational fluid dynamics, Reto Fiolka completed his PhD at the institute of Nanotechnology at ETH Zurich in the group of Dr Andreas Stemmer. He conducted post-doctoral research at the Howard Hughes Medical Institute's Janelia Research Campus under the late Dr Mats Gustafsson working on 3D structured illumination microscopy and under Dr Meng Cui on adaptive optics. At UT Southwestern, the research in his lab aims to extend the current imaging capabilities of optical microscopy such that cancer cell research and drug screening can be performed in physiologically relevant, 3D environments, ex vivo and in vivo. His microscope development is focused on improving the spatiotemporal resolution and optical penetration depth and translating new technologies to biological research. |
|---|---|
| 14:00 - 14:30 |
High-speed and cost-efficient super-resolution structured illumination microscopy enabled entirely by fiber optics
Super-resolved structured illumination microscopy (SR-SIM) is among the most flexible, fastest and least perturbing fluorescence microscopy techniques capable of surpassing the optical diffraction limit. Current custom-built instruments are easily able to deliver two-fold resolution enhancement at video-rate frame rates, but the cost of the instruments is still relatively high and the physical size of the instruments is still prohibitively large. Here, Professor Huser will present his and his team's latest efforts towards realising a new generation of compact, cost-efficient and high-speed SR-SIM instruments based entirely on fiber-optics. He will discuss the technical approaches which we have taken in order to realize a highly robust from of 2D- and TIRF-based SR-SIM, and show applications of cell biological data obtained with this instrument. He will also discuss the influence of the modulation transfer function of different types of sCMOS cameras on the overall system performance, in particular the spatial resolution that can be obtained with such an instrument. 
Professor Thomas Huser, University of Bielefeld, Germany

Professor Thomas Huser, University of Bielefeld, GermanyProfessor Thomas Huser is a professor of Physics in the Department of Physics at the University of Bielefeld, Germany. He has been a university professor for more than 15 years and a scientific group leader for the last 20 years. Professor Huser started his scientific career as a postdoctoral fellow at the Lawrence Livermore National Laboratory in Livermore, California, where he became scientific capability leader for Nano-Biophotonics and became an Associated Professor at the University of California, Davis, in 2005, where he also acted as Chief Scientist for the NSF Center for Biophotonics. From 2009 - 2011 he was also a guest professor at the University of Tromsø in Norway. His research interests are ultrasensitive optical microscopy and spectroscopy at the level of single cells and single molecules, applied to biomedical problems with high relevance for human health. |
| 14:30 - 15:00 |
Multi-SIM via deep learning algorithm for super-resolution live imaging
Biology research is desired to characterise the intracellular dynamics at high spatiotemporal resolution with low photobleaching and phototoxicity effects, which therefore allow continuously resolve the delicate structures and behaviors of the engaged organelles over the whole biological process. However, the trade-offs between spatial and temporal resolution, and low phototoxicity/photobleaching always compromise the practical performance of current super-resolution imaging techniques. To achieve these normally opposing goals, in this talk Professor Dong Li will discuss his and his team's latest developments in multi-modality structured illumination microscopy (Multi-SIM) and lattice light sheet SIM microscopy (LLS-SIM), which enable high-speed super-resolution live-cell imaging for thousands of time-points spanning over hours of time-lapse. Recently, they further developed a deep-learning algorithm for super-resolution image reconstruction, termed deep Fourier channel attention network (DFCAN), which further extends the applicability of Multi-SIM and LLS-SIM into more challenging imaging conditions. 
Professor Dong Li, Institute of Biophysics, Chinese Academy of Sciences, China

Professor Dong Li, Institute of Biophysics, Chinese Academy of Sciences, ChinaDr Dong Li is Principal Investigator at Institute of Biophysics. He received his PhD in electronic and computer engineering at Hong Kong University of Science and Technology in 2011. Then he spent four years for postdoctoral research of developing super-resolution microscopy at Janelia Research Campus, Howard Hughes Medical Institute. After that he joined Institute of Biophysics. Dr Dong Li’s group focuses on developing super-resolution microscopy (SIM) for high-speed and long-term live imaging, and exploring their applications in biological research. Recently, Dong has made advances in patterned activation nonlinear SIM (PA NL-SIM) that breaks the 100 nm resolution barrier of conventional SIM for live-cell imaging; grazing incidence SIM (GI-SIM) that optimizes the 2D illumination depth while suppressing out-of-focus background; deep Fourier channel attention network SIM (DFCAN-SIM) that achieved robust SIM reconstruction at the challenging imaging conditions. These techniques enable interrogate the dynamics of biological processes in live status at ultrahigh resolution in space and time. |
| 15:00 - 15:30 | Tea |
| 15:30 - 16:00 |
Home-built SIM with SLM and Multifocus Microscopy
The team's multifocus structured illumination microscope (MF-SIM) for high-speed super-resolution microscopy in 3D is currently being launched in the UCSC imaging facility. They here describe this new instrument and their solutions to the hardware challenges in terms of optics, opto-mechanics and electronics using a liquid crystal spatial light modulator to generate the SIM pattern with respect to optical design and acquisition timing. 
Dr Sara Abrahamsson, University of California Santa Cruz, USA

Dr Sara Abrahamsson, University of California Santa Cruz, USASara Abrahamsson is an Assistant Professor at UCSC pursuing groundbreaking research in optical systems design and diffractive optics for 3D microscopy and super-resolution microscopy. She has developed the imaging methods aberration-corrected Multifocus Microscopy and MultiFocus Structured Illumination Microscopy which enable live simultaneous 3D imaging at and beyond the diffraction limit of resolution. 
Mr Eduardo Hirata, University of California Santa Cruz, USA

Mr Eduardo Hirata, University of California Santa Cruz, USAEduardo Hirata is a PhD candidate in the SaraLab and is in the final stages of launching his 25-camera Multifocus Microscope (M25) which is employed in high-speed functional neuronal imaging. |
| 16:00 - 16:20 |
Digital micromirror devices for cost-effective and fast multi-color structured illumination microscopy
Modern structured illumination microscopes (SIMs) often rely on spatial light modulators (SLMs) to allow for a fast and robust implementation of the SIM technique. Of the different SLM technologies available, digital micromirror devices (DMDs) feature many advantages, such as high speed, low cost, and optical properties such as wavelength range and polarisation. However, they also pose a challenge, as their jagged surface introduces a blazed grating effect, that has to be carefully accounted for when using these devices with the coherent light sources needed for SIM. In this talk, both a simulation framework and the experimental implementations of DMDs for the use in SIM will be discussed. The simulation framework is used to find wavelength combinations suitable for multi-color DMD-based SIM, and to assess the systems alignment parameters. An experimental implementation showcases how these results can be used to create a fast, robust and very cost-effective dual- and multi-color DMD-SIM system. 
Dr Marcel Müller, Bielefeld University, Germany

Dr Marcel Müller, Bielefeld University, GermanyMarcel Müller works on implementing microscope systems and data reconstruction algorithms for structured illumination microscopy (SIM). In 2016, he started the fairSIM project, which aims to provide SIM-related resources (software plugins, example datasets, system blueprints) to the scientific community. He has since worked at the Micron Advanced Imaging Facility at the University of Oxford, as a MSCA fellow at the KU Leuven, and is currently a Postdoctoral Researcher at Bielefeld University in Germany. His research focused on new, robust, fast and cost-effective implementations of structured illumination microscopy. |
| 16:20 - 16:40 |
GPU-accelerated real-time reconstruction in Python of three-dimensional datasets from structured illumination microscopy with hexagonal patterns
The researchers present a structured illumination microscopy system that projects a hexagonal pattern by the interference among three coherent beams, suitable for implementation in a light-sheet geometry. Seven images acquired as the illumination pattern is shifted laterally can be processed to produce a super-resolved image that surpasses the diffraction-limited resolution by a factor of over 2 in an exemplar light-sheet arrangement. Three methods of processing data are discussed depending on whether the raw images are available in groups of seven, individually in a stream or as a larger batch representing a three-dimensional stack. The researchers show that imaging axially moving samples can introduce artefacts, visible as fine structures in the processed images. However, these artefacts are easily removed by a filtering operation carried out as part of the batch processing algorithm for three- dimensional stacks. The reconstruction algorithms implemented in Python include specific optimisations for calculation on a graphics processing unit and we demonstrate its operation on experimental data of static objects and on simulated data of moving objects. They show that the software can process over 239 input raw frames per second at 512×512 pixels, generating over 34 super-resolved frames per second at 1024×1024 pixels. 
Dr Hai Gong, Imperial College London, UK

Dr Hai Gong, Imperial College London, UKHai Gong received the BS degree in Mechatronic Engineering from Harbin Institute of Technology in 2007, the MS degree in Mechanical Engineering from Xi’an Jiaotong University in 2011, and the PhD degree in optics and control engineering from Delft University of Technology in 2019. He is currently working as a postdoctoral Research Associate in the Department of Physics at Imperial College London. His research interests include structured illumination microscopy, light-sheet microscopy, wavefront sensing and adaptive optics. In 2017, he received the silver Edmund Optics educational award. |
| 16:40 - 17:00 |
mmSIM: an open toolbox for accessible structured illumination microscopy
Over the past two decades structured illumination microscopy (SIM) has proven to be a powerful method for high-speed fluorescence imaging beyond the classical diffraction limit. This presentation will describe a simple open source approach, termed mmSIM, for controlling SIM hardware and acquiring SIM images based on the popular MicroManager software package. By complementing existing hardware designs and open source image reconstruction software, mmSIM supports the keen microscopist to develop and run their own custom-built SIM system. The configuration and performance of two mmSIM-controlled spatial light modulator-based SIM systems will discussed along with results from various bioimaging studies performed using the devices, including measurement of the kinetics of in vitro protein fibrillogenesis and visualisation of intracellular uptake. 
Dr Michael Shaw, National Physical Laboratory and Department of Computer Science, University College London, UK

Dr Michael Shaw, National Physical Laboratory and Department of Computer Science, University College London, UKMike joined the National Physical Laboratory (NPL) after completing an undergraduate degree in physics at Imperial College London. He completed his PhD studies, also with Imperial College, on overcoming resolution limits in fluorescence microscopy using adaptive optics and structured illumination whilst working at NPL. Mike is currently a Principal Research Scientist in NPL’s Biometrology group where his work focuses on the development and application of high-resolution fluorescence microscopy techniques for multiscale biological imaging, particularly structured illumination and light sheet methods. He is also a Principal Research Fellow in the Department of Computer Science at University College London, where he leads a multidisciplinary team based in the UCL TouchLab exploring label free computational imaging, image analysis and manipulation techniques for diagnostic and therapeutic applications. 
Dr Craig Russell, EMBL-EBI, UK

Dr Craig Russell, EMBL-EBI, UKCraig is a Computational Microscopist who develops software and algorithms for addressing microscopy related challenges. During his PhD he built a light-sheet microscope for developmental biology; he then spent some time at NPL working, developing SIM and airy-light-sheet systems for BioMetrology; and he now works as a Data Scientist at the EMBL-EBI using machine learning for analysing biological images. |
| 17:00 - 17:45 |
Panel discussion: Biological application of SIM - from experimental design to quantitative evaluation

Dr Lothar Schermelleh, University of Oxford, UK

Dr Lothar Schermelleh, University of Oxford, UKLothar Schermelleh has studied Biology at the Ludwig Maximilian University Munich (LMU). He obtained his doctorate under the supervision of Thomas Cremer in 2003 for his work on the 'Dynamic organisation of chromosomes in the mammalian cell nucleus'. He later joined the group of Heinrich Leonhardt as a Postdoctoral Researcher and Lecturer, where he studied the role and regulation of DNA methylation 1 through developing and applying advanced single-cell imaging and analyses approaches. During this time, as a visiting scientist in the lab of John Sedat at the UCSF, he first came in contact with super-resolution structured illumination microscopy (SIM), and he later established this technique and its cell biological application at the LMU Munich. In 2011, Lothar joined the University of Oxford as Micron Senior Research Fellow and Principal Investigator at the Department of Biochemistry, where his group further advanced the development of quantitative super-resolution 3D imaging to study mesoscale chromatin organisation and mechanisms of epigenetic gene regulation, eg in X chromosome inactivation. In 2020, he was named Associate Professor and became Academic Director of the Micron Bioimaging Facility. 
Professor Thomas Huser, University of Bielefeld, Germany

Professor Thomas Huser, University of Bielefeld, GermanyProfessor Thomas Huser is a professor of Physics in the Department of Physics at the University of Bielefeld, Germany. He has been a university professor for more than 15 years and a scientific group leader for the last 20 years. Professor Huser started his scientific career as a postdoctoral fellow at the Lawrence Livermore National Laboratory in Livermore, California, where he became scientific capability leader for Nano-Biophotonics and became an Associated Professor at the University of California, Davis, in 2005, where he also acted as Chief Scientist for the NSF Center for Biophotonics. From 2009 - 2011 he was also a guest professor at the University of Tromsø in Norway. His research interests are ultrasensitive optical microscopy and spectroscopy at the level of single cells and single molecules, applied to biomedical problems with high relevance for human health. 
Dr Liangyi Chen, Peking University, China

Dr Liangyi Chen, Peking University, ChinaLiangyi Chen is Boya Professor of Peking University. He obtained his undergraduate degrees Biomedical engineering in Xi’an JiaoTong University, then majored in Biomedical engineering in pursuing PhD degree in Huazhong University of Science and Technology. His lab focused on two interweaved aspects: the development of new imaging and quantitative image analysis algorithms, and the application of these technology to study how glucose-stimulated insulin secretion is regulated in the health and disease at multiple levels (single cells, islets and in vivo) in the health and disease animal models. The techniques developed included ultrasensitive Hessian structured illumination microscopy (Hessian SIM) for live cell super-resolution imaging, the Sparse deconvolution algorithm for extending spatial resolution of fluorescence microscopes limited by the optics, Super-resolution fluorescence-assisted diffraction computational tomography (SR-FACT) for revealing the three-dimensional landscape of the cellular organelle interactome, two-photon three-axis digital scanned lightsheet microscopy (2P3A-DSLM) for tissue and small organism imaging, and fast High-resolution Miniature Two-photon Microscopy (FHIRM-TPM) for Brain Imaging in Freely-behaving Mice. He is also recipient of the National Distinguish Scholar Fund project from National Natural Science Foundation of China. 
Dr Michael Shaw, National Physical Laboratory and Department of Computer Science, University College London, UK

Dr Michael Shaw, National Physical Laboratory and Department of Computer Science, University College London, UKMike joined the National Physical Laboratory (NPL) after completing an undergraduate degree in physics at Imperial College London. He completed his PhD studies, also with Imperial College, on overcoming resolution limits in fluorescence microscopy using adaptive optics and structured illumination whilst working at NPL. Mike is currently a Principal Research Scientist in NPL’s Biometrology group where his work focuses on the development and application of high-resolution fluorescence microscopy techniques for multiscale biological imaging, particularly structured illumination and light sheet methods. He is also a Principal Research Fellow in the Department of Computer Science at University College London, where he leads a multidisciplinary team based in the UCL TouchLab exploring label free computational imaging, image analysis and manipulation techniques for diagnostic and therapeutic applications. 
Professor Dong Li, Institute of Biophysics, Chinese Academy of Sciences, China

Professor Dong Li, Institute of Biophysics, Chinese Academy of Sciences, ChinaDr Dong Li is Principal Investigator at Institute of Biophysics. He received his PhD in electronic and computer engineering at Hong Kong University of Science and Technology in 2011. Then he spent four years for postdoctoral research of developing super-resolution microscopy at Janelia Research Campus, Howard Hughes Medical Institute. After that he joined Institute of Biophysics. Dr Dong Li’s group focuses on developing super-resolution microscopy (SIM) for high-speed and long-term live imaging, and exploring their applications in biological research. Recently, Dong has made advances in patterned activation nonlinear SIM (PA NL-SIM) that breaks the 100 nm resolution barrier of conventional SIM for live-cell imaging; grazing incidence SIM (GI-SIM) that optimizes the 2D illumination depth while suppressing out-of-focus background; deep Fourier channel attention network SIM (DFCAN-SIM) that achieved robust SIM reconstruction at the challenging imaging conditions. These techniques enable interrogate the dynamics of biological processes in live status at ultrahigh resolution in space and time. 
Dr Jennifer Lippincott-Schwartz, Janelia Research Campus, USA

Dr Jennifer Lippincott-Schwartz, Janelia Research Campus, USADr Jennifer Lippincott-Schwartz is a Senior Group Leader at the Howard Hughes Medical Institute’s Janelia Research Campus. She has pioneered the use of green fluorescent protein technology for quantitative analysis and modelling of intracellular protein traffic and organelle dynamics in live cells. Her innovative techniques to label, image, quantify and model specific live cell protein populations and track their fate have provided vital tools used throughout the research community. Her own findings using these techniques have reshaped thinking about the biogenesis, function, targeting, and maintenance of various subcellular organelles and macromolecular complexes and their crosstalk with regulators of the cell cycle, metabolism, aging, and cell fate determination. She is an elected member of the National Academy of Sciences, the National Academy of Medicine, the American Society of Arts and Sciences and the European Molecular Biology Organization. She is also a Fellow of The Biophysical Society, The Royal Microscopical Society and The American Society of Cell Biology. Her awards include the E.B. Wilson Medal of the American Society of Cell Biology, the Newcomb Cleveland Prize of the American Association for the Advancement of Science, the Van Deenen Medal, the Keith Porter Award of the American Society of Cell Biology, the Feodor Lynen Medal, and the Feulgen Prize of the Society of Histochemistry. She co-authored of the textbook Cell Biology and was President of the American Society of Cell Biology. Dr Lippincott-Schwartz attended Swarthmore College, received her MS from Stanford University, and obtained her PhD in Biochemistry from Johns Hopkins University in 1986. |
Chair

Dr Benedict Diederich, Leibniz Institute of Photonic Technology, Germany

Dr Benedict Diederich, Leibniz Institute of Photonic Technology, Germany
After doing an apprenticeship as an electrician, Benedict Diederich started studying electrical engineering at the University for Applied Science Cologne. A specialization in optics and an internship at Nikon Microscopy Japan pointed him to the interdisciplinary field of microscopy. After working for Zeiss, he started his PhD in the Heintzmann Lab at the Leibniz IPHT Jena. He focuses on bringing cutting-edge research to everybody by relying on tailored image processing and low-cost optical setups. Part of his PhD program took place at the Photonics Centre at Boston University in the Tian Lab. A recent contribution was the open-source optical toolbox UC2 (You-See-Too) which tries to democratize science by making cutting-edge affordable and available to everyone, everywhere. He is co-founder of the open-hardware start-up “openUC2” which aims to scale up open microscopy to solve global problems and spend some time during his Post Doc in Manu Prakash’s lab at Stanford University to promote “Frugal Optics” in marine biology communities.
| 09:00 - 09:30 |
Keynote: Modulated excitation for enhanced single-molecule localisation microscopy
In Single Molecule Localization Microscopy, the localisation precision relies strongly on the spatial analysis of the point spread function, which quickly degrades with increasing depth due to aberrations, impacting both lateral and axial resolution. Alternative localisation strategies have been proposed using time varying structured illumination based on traveling interferences or more recently on triangulation from a zero-intensity point of the excitation beam. These strategies achieve a more precise localisation precision with less photons, but are designed for single point analysis and thus required scanning. This concept has been revisited in wide field approach with different implementation to enhanced lateral or axial resolution. Dr Lévêque-Fort will present hers and her team's strategy based on the modulation of the fluorescence emission using a periodically wide field structured excitation, called ModLoc for Modulation Localization. The position of a fluorescent molecule within the moving fringe pattern is encoded in the phase of its modulated emission signal. The camera being slow, the signal demodulation is performed by a specific optical assembly placed in front of the camera. Dr Lévêque-Fort will show performances on ModLoc for lateral precision but also for 3D imaging where a uniform axial precision of ~6.8 nm can be reached, and imaging at 50 µm in depth obtained. 
Dr Sandrine Lévêque-Fort, CNRS, Université Paris Saclay, France

Dr Sandrine Lévêque-Fort, CNRS, Université Paris Saclay, FranceSandrine Lévêque-Fort is a CNRS Researcher Director at the Institute of molecular science (ISMO) in Paris Saclay University. She obtained her PhD on the development of a new acousto-optic imaging approach for imaging through scattering media in the Optical Lab of ESPCI in Paris. She then became a postdoctoral fellow in the physics department of Imperial College, where she started to develop time resolved fluorescence microscopy but also structured illumination strategy. She joined the CNRS in 2001 to develop different strategies to improve spatial and temporal resolution for fluorescence microscopy, by implementing new configurations or developed plasmonics substrates to engineered fluorescence emission. Since 2009, she has proposed various approaches to take advantage of supercritical angle fluorescence (SAF) emission as an alternative intrinsic tool given by the fluorophore itself to access axial information, in association with classical and super-resolution microscopy. Since 2016, she has combined structured excitation with single molecule localisation. By introducing a time signature within the localisation process, this technique called ModLoc permits to retrieve the fluorophores’ information thanks to the phase of their modulated emission and benefits of an enhanced localisation. |
|---|---|
| 09:30 - 10:00 |
Spatially Modulated Illumination Microscopy: Application Perspectives in nuclear Nanostructure Analysis
The spatial organisation of the cell nucleus of higher organisms has emerged as a main topic of advanced light microscopy. So far, a variety of super-resolution methods have been applied for this, including 4Pi-, STED-, and Localisation Microscopy approaches, as well as various approaches of Structured Illumination Microscopy. Here we summarise the state of the art and discuss application perspectives for nuclear nanostructure analysis of Spatially Modulated Illumination (SMI). SMI is a widefield based approach to use axially structured illumination patterns to determine the extension (size) of small, optically isolated fluorescent objects between ≤ 200 nm and ≥ 40 nm diameter with a precision down to the few nm range; in addition, it allows the axial positioning of such structures down to the 1 nm scale; combined with laterally structured illumination, a 3D localisation precision of ≤1 nm is expected to become feasible using fluorescence yields typical for Single Molecule Localisation Microscopy (SMLM) applications. Together with its nanosising capability, this may eventually be used to analyse macromolecular complexes and other nanostructures with a topological resolution further narrowing the gap to Cryoelectron microscopy. 
Professor Christoph Cremer, Kirchhoff Institute for Physics, University Heidelberg and Max-Planck Institute for Polymer Physics, Germany

Professor Christoph Cremer, Kirchhoff Institute for Physics, University Heidelberg and Max-Planck Institute for Polymer Physics, GermanyChristoph Cremer studied Physics at the Universities of Freiburg and Munich (Germany) and obtained a Dr.rer.nat. (PhD) in Biophysics and Genetics at University of Freiburg, as well as a degree in General Human Genetics & Experimental Cytogenetics. In 1983 he was appointed Professor of Applied Optics & Information Processing at Heidelberg University. In addition, he is an Honorary Professor (Physics) at the Johannes Gutenberg University Mainz, Germany, and a Research Associate at the Max Planck Institutes for Polymer Research, and for Chemistry (Mainz), respectively. The main goal of his research is to develop and apply advanced light microscopy methods to elucidate biological nanostructures. A special focus is given to the cell nucleus as the seat of genetic information and of gene regulation. Presently an intranuclear optical resolution down to ca. 5 nm (1/100th of the exciting wavelength) has been achieved. |
| 10:00 - 10:30 |
At the molecular resolution with MINFLUX?
MINFLUX is a promising new development in single-molecule localisation microscopy, claiming a resolution of 1-3 nm in living and fixed biological specimens. While MINFLUX can achieve very high localisation precision, quantitative analysis of reported results leads us to dispute the resolution claim and question reliability for imaging sub-100-nm structural features, in its current state. 
Dr Kirti Prakash, The Institute of Cancer Research, UK.

Dr Kirti Prakash, The Institute of Cancer Research, UK.Kirti Prakash is a computer scientist by training (Bachelors and Masters degree) but a biologist at heart (PhD degree). Kirti aspires to be an inventor and develop new imaging tools for cell biology and neuroscience. Kirti did his Masters in Computer Science from Aalto University (Finland) and PhD in Biology from Heidelberg University (Germany). During his PhD, he developed a new method to image DNA which led to the first high-resolution images of the epigenetic landscape of meiotic chromosomes and mechanisms behind chromosome condensation. The doctoral research earned him several awards including Springer Best PhD Thesis Prize. After his PhD, he did a couple of postdocs at Carnegie Institution for Science (USA) and University of Cambridge (UK). The primary highlights of his research here were laser-free superresolution microscopy and the development of a high-content imaging pipeline to quantify single-cell gene expression. He then spent a couple of years at the National Physical Laboratory (UK) to work on standardisation and quality control of quantitative microscopy. Presently at the Institute of Cancer Research (UK), he is working on artificial intelligence and digital pathology. |
| 10:30 - 11:00 | Coffee |
| 11:00 - 11:20 |
Upscaling SIM reconstruction via Deep Learning
Structured Illumination Microscopy (SIM) is a widespread methodology to image live and fixed biological structures smaller than the diffraction limits of conventional optical microscopy. Using recent advances in image up-scaling through deep learning models, we demonstrate a method to reconstruct 3D SIM image stacks with twice the axial resolution attainable through conventional SIM reconstructions. We further demonstrate our method is robust to noise and evaluate it against two-point cases and axial gratings. Finally, we discuss potential adaptions of the method to further improve resolution. 
Mr Miguel Boland, Imperial College London, UK

Mr Miguel Boland, Imperial College London, UKMiguel is a Wellcome Trust PhD student in Mathematics at Imperial College, London. During his Masters at Imperial, he worked under the guidance of Professor Mark Neil, Dr Ed Cohen and Dr Seth Flaxman to develop an upscaling model applied to the reconstruction of SIM images. Miguel has previously worked at the European Bioinformatics Institute in Cambridge, where he contributed to research in metagenomics by implementing data production pipelines and websites for the meta-analysis and sharing of microbial genomic datasets. His PhD now focuses on applications of Deep Learning in SMLM microscopy, and more particularly in 3D molecule localisation and clustering algorithms using Graph Neural Networks. |
| 11:20 - 11:40 |
Polarised Illumination Coded Structured Illumination Microscopy (picoSIM): Experimental Results
The need for acquiring at least three images to reconstruct an optical section of a sample limits the acquisition rate in structured illumination microscopy (SIM) for optical sectioning. In polarised illumination coded structured illumination microscopy (picoSIM) the three illumination patterns are encoded in a single polarised illumination light distribution. This distribution consists of linearly polarised light, with the polarisation orientation varying with the position in the focal plane. If the sample exhibits sufficient fluorescence anisotropy, this linear polarisation will still be present to a certain degree in the emitted light. Splitting the emission light and filtering the different parts with polarisation analysers of differing orientation thus allows the acquisition of the complete SIM data in a single exposure. In this presentation Dr Wicker describes the theoretical background of picoSIM and presents an experimental set-up and first experimental results. 
Dr Kai Wicker, ZEISS Innovation Hub Dresden, Germany

Dr Kai Wicker, ZEISS Innovation Hub Dresden, GermanyKai is a physicist with a background in quantum optics and information. In 2006 he changed tack to do his PhD in the group of Rainer Heintzmann at King’s College London, where he worked on the development of various microscopy techniques, a major focus being reconstruction algorithms for SIM. After a PostDoc at University of Jena, again in the Heintzmann group, he joined ZEISS’ corporate research department in 2013, where he has held various roles. He is currently heading the new ZEISS Innovation Hub at Technische Universität Dresden, driving co-innovation between industry and academia. |
| 11:40 - 12:00 |
Modular Microscopy – not just a Toy
Modern microscopy methods enable impressive images that provide deeper insights for interdisciplinary research. Very often, however, these methods are accompanied by complex optical setups, which leads to a high price and unfortunately gives these methods a certain exclusivity. With UC2 (You-See-Too) the researchers have recently introduced a low-cost modular 3D printable microscopy toolkit that aims to create an open standard in optics. It simplifies the creation and sharing of arbitrarily complicated optical setups. The growing community of educators, developers and users established easy-to-use systems, such as the light sheet microscope or the 'openSIM' for structured illumination with microscopic super resolution. UC2 can help biologists to answer new questions with highly available microscopes or support university students to understand the basic principles of optics by challenging their creativity. UC2 helps bringing different disciplines closer together, since different strategies can easily be discussed and jointly implemented within a project – even 'over Zoom'. Dr Diederich presents a series of advances the modular open-source toolbox acquired since it was first introduced. This is the result from user feedback and a long-lasting learning curve, where iterative design strategies helped to build a product-like research project. By developing cost-effective instruments and sharing designs and manuals, the stage is set for the democratisation of super-resolution imaging. 
Dr Benedict Diederich, Leibniz Institute of Photonic Technology, Germany

Dr Benedict Diederich, Leibniz Institute of Photonic Technology, GermanyAfter doing an apprenticeship as an electrician, Benedict Diederich started studying electrical engineering at the University for Applied Science Cologne. A specialization in optics and an internship at Nikon Microscopy Japan pointed him to the interdisciplinary field of microscopy. After working for Zeiss, he started his PhD in the Heintzmann Lab at the Leibniz IPHT Jena. He focuses on bringing cutting-edge research to everybody by relying on tailored image processing and low-cost optical setups. Part of his PhD program took place at the Photonics Centre at Boston University in the Tian Lab. A recent contribution was the open-source optical toolbox UC2 (You-See-Too) which tries to democratize science by making cutting-edge affordable and available to everyone, everywhere. He is co-founder of the open-hardware start-up “openUC2” which aims to scale up open microscopy to solve global problems and spend some time during his Post Doc in Manu Prakash’s lab at Stanford University to promote “Frugal Optics” in marine biology communities. |
| 12:00 - 12:45 |
Workshop: UC2 - an open source system for optics (The power in your pocket)
With UC2 [1] the researchers from the Leibniz-IPHT in Jena are aiming for nothing less than a revolution in optics. Just as the Arduino made electronics and microcontroller programming more accessible outside the field, the open source optics toolbox aims to do the same, but for optics. The workshop will give you a comprehensive introduction to the documentation and guide you through the process of creating your first setup. Based on this knowledge, the researchers introduce the modular developer kit (MDK, [2]), which helps you to bring in your own ideas and parts. The workshop is meant to serve as a starting point for people who are new to 3D printing in optics and microscopy in general and UC2 in particular. Besides that, the researchers will present recent advances of UC2 setups like the galvo-based light sheet microscope or the in vitro fluorescence microscope that can image multiple fluorescent dyes, and show how important open source software and hardware projects are to promote reproducible research 'out of the box'. [1] Diederich, B, Lachmann, R, Carlstedt, S et al A versatile and customizable low-cost 3D-printed open standard for microscopic imaging. Nat Commun 11, 5979 (2020). [2] UC2 GitHub Repository: https://github.com/openuc2/UC2-GIT

Dr Benedict Diederich, Leibniz Institute of Photonic Technology, Germany

Dr Benedict Diederich, Leibniz Institute of Photonic Technology, GermanyAfter doing an apprenticeship as an electrician, Benedict Diederich started studying electrical engineering at the University for Applied Science Cologne. A specialization in optics and an internship at Nikon Microscopy Japan pointed him to the interdisciplinary field of microscopy. After working for Zeiss, he started his PhD in the Heintzmann Lab at the Leibniz IPHT Jena. He focuses on bringing cutting-edge research to everybody by relying on tailored image processing and low-cost optical setups. Part of his PhD program took place at the Photonics Centre at Boston University in the Tian Lab. A recent contribution was the open-source optical toolbox UC2 (You-See-Too) which tries to democratize science by making cutting-edge affordable and available to everyone, everywhere. He is co-founder of the open-hardware start-up “openUC2” which aims to scale up open microscopy to solve global problems and spend some time during his Post Doc in Manu Prakash’s lab at Stanford University to promote “Frugal Optics” in marine biology communities. 
Mr Haoran Wang, Leibniz-Institut für Photonische Technologien, Germany

Mr Haoran Wang, Leibniz-Institut für Photonische Technologien, GermanyHaoran Wang received the MSc degree in the Leibniz University Hannover, Germany, in 2020, with the topic of fibre-based optical manipulation with a 3D-printed microfluidic chip. He is currently a PhD student in the Heintzmann Lab at the Leibniz-Institute of Photonic Technology, Jena, Germany. His research focus is structured illumination microscopy and frugal science using cellphones. |
Chair

Dr Stefanie Reichelt, University of Cambridge, UK

Dr Stefanie Reichelt, University of Cambridge, UK
Stefanie established and led the light microscopy core at the Cancer Research UK Cambridge Institute (CRUK CI) for 16 years, which included the development of new imaging techniques enabling the visualisation of molecules in cells for cancer diagnostics. Prior to this, Stefanie contributed to the development and commercialisation of laser scanning microscopy with Brad Amos, FRS, at the MRC-LMB and Bio-Rad Microscience. She is the co-founder of the Plymouth Advanced Microscopy Course and has established the Cambridge Technology Platforms Network (CTPN) and the Royal Microscopical Society Imaging One World lecture series. In her current role as Public Engagement Manager for the Biomedical schools at the University of Cambridge, UK, Stefanie contributes to the public understanding of science.
| 13:30 - 14:00 |
Keynote: Looking under the hood of cells: from single molecule dynamics to whole cell organelle reconstructions
Powerful new ways to image the internal structures and complex dynamics of cells are revolutionising cell biology and bio-medical research. In this talk, Dr Lippincott-Schwartz will focus on how emerging fluorescent technologies are increasing spatio-temporal resolution dramatically, permitting simultaneous multispectral imaging of multiple cellular components. In addition, results will be discussed from whole cell milling using Focused Ion Beam Electron Microscopy (FIB-SEM), which reconstructs the entire cell volume at 4 voxel resolution. Using these tools, it is now possible to begin constructing an 'organelle interactome', describing the interrelationships of different cellular organelles as they carry out critical functions. The same tools are also revealing new properties of organelles and their trafficking pathways, and how disruptions of their normal functions due to genetic mutations may contribute to important diseases. 
Dr Jennifer Lippincott-Schwartz, Janelia Research Campus, USA

Dr Jennifer Lippincott-Schwartz, Janelia Research Campus, USADr Jennifer Lippincott-Schwartz is a Senior Group Leader at the Howard Hughes Medical Institute’s Janelia Research Campus. She has pioneered the use of green fluorescent protein technology for quantitative analysis and modelling of intracellular protein traffic and organelle dynamics in live cells. Her innovative techniques to label, image, quantify and model specific live cell protein populations and track their fate have provided vital tools used throughout the research community. Her own findings using these techniques have reshaped thinking about the biogenesis, function, targeting, and maintenance of various subcellular organelles and macromolecular complexes and their crosstalk with regulators of the cell cycle, metabolism, aging, and cell fate determination. She is an elected member of the National Academy of Sciences, the National Academy of Medicine, the American Society of Arts and Sciences and the European Molecular Biology Organization. She is also a Fellow of The Biophysical Society, The Royal Microscopical Society and The American Society of Cell Biology. Her awards include the E.B. Wilson Medal of the American Society of Cell Biology, the Newcomb Cleveland Prize of the American Association for the Advancement of Science, the Van Deenen Medal, the Keith Porter Award of the American Society of Cell Biology, the Feodor Lynen Medal, and the Feulgen Prize of the Society of Histochemistry. She co-authored of the textbook Cell Biology and was President of the American Society of Cell Biology. Dr Lippincott-Schwartz attended Swarthmore College, received her MS from Stanford University, and obtained her PhD in Biochemistry from Johns Hopkins University in 1986. |
|---|---|
| 14:00 - 14:20 |
Structured illumination ophthalmoscope: super-resolution microscopy on the living human eye
Here, the researchers present the prototype of an ophthalmoscope that uses structured illumination microscopy (SIM) to enable super-resolved imaging of the human retina and give first insights into clinical application possibilities. The SIM technique was applied to build a prototype that uses the lens of the human eye as an objective to ‘super-resolve’ the retina of a living human. In their multidisciplinary collaboration, the researchers have adapted this well-established technique to ophthalmology and successfully imaged a human retina using significantly lower light intensity than a state of the art ophthalmoscope. They focus on the technical implementation and highlight future perspectives of this method. 
Florian Schock, Heildelberg University, Germany

Florian Schock, Heildelberg University, Germany |
| 14:20 - 14:40 |
Extended mechanical force measurements using structured illumination microscopy
Quantifying cell generated mechanical forces is key to furthering our understanding of mechanobiology. Traction force microscopy (TFM) is one of the most broadly applied force probing technologies, but its sensitivity is strictly dependent on the spatio-temporal resolution of the underlying imaging system. In previous works, it was demonstrated that increased sampling densities of cell derived forces permitted by super-resolution fluorescence imaging enhanced the sensitivity of the TFM method. However, these recent advances to TFM based on super-resolution techniques were limited to slow acquisition speeds and high illumination powers. Here, the researchers present three novel TFM approaches that, in combination with total internal reflection, structured illumination microscopy and astigmatism, improve the spatial and temporal performance in either two-dimensional or three-dimensional mechanical force quantification, while maintaining low illumination powers. These three techniques can be straightforwardly implemented on a single optical set-up offering a powerful platform to provide new insights into the physiological force generation in a wide range of biological studies. 
Dr Kseniya Korobchevskaya, University of Oxford, UK

Dr Kseniya Korobchevskaya, University of Oxford, UKKseniya received her MSc degree from Novosibirsk State Technical University, Russia, in the field of physics of optical effects. After she has joined the Italian Institute of Technology (IIT), Italy, for PhD in the topic of femtosecond time resolved spectroscopy. Currently Kseniya is a member of Biophysical Immunology group at Kennedy Institute of Rheumatology (KIR), Oxford. Her research is focused on development and application of advance and super-resolution microscopy techniques for studying the sub-cellular structures and biological processes during immune cell interactions. |
| 14:40 - 15:00 |
Quality control of image sensors using Gaseous Tritium Light Sources
Cameras and other detectors are indispensable tools for modern light microscopy. Performance may, however, vary from device to device and they can show signs of damage or ageing which can, in turn, affect the quality and reproducibility of data. Despite this, many labs do not regularly perform quantitative quality control checks on their instruments. One explanation could be a relative lack of convenient and low-cost calibration sources. Work by David McFadden, Brad Amos and Rainer Heintzmann proposes to tackle this problem using inexpensive tritium radioluminescent tubes (betalights). The mechanical design is easily reproducible and can be 3D-printed. Another major advantage of the design is that the calibration allows for a plug and play approach with automatic image analysis based on the photon transfer method. The calibration yields results for the photon conversion factor and read noise as well as the detector quantum efficiency. The intensity is suitable for calibrating detectors at very low light levels, characteristic especially of single-molecule-localisation microscopy. 
Mr David McFadden, Friedrich-Schiller-Universität Jena, Germany

Mr David McFadden, Friedrich-Schiller-Universität Jena, GermanyDavid McFadden is a PhD student at the Friedrich Schiller University Jena and working in the microscopy group at the Leibniz Institute of Photonic Technology. His current work aims to understand the limitations that detectors impose upon novel methods such as super-resolution microscopy and quantum-enhanced detection schemes. |
| 15:00 - 15:30 | Tea |
| 15:30 - 15:50 |
Laser-free super-resolution microscopy
A new single-molecule localisation microscopy (SMLM) configuration termed laser-free super-resolution microscopy (LFSM) is presented. LFSM enables high-density single-molecule super-resolution microscopy with a conventional epifluorescence microscope set-up and a mercury arc lamp. The setup allows single molecules to be switched on and off (a phenomenon termed as ‘blinking’), detected and localised. The use of a short burst of deep blue excitation (350–380 nm) can be further used to reactivate the blinking, once the blinking process has slowed or stopped. A resolution of 90 nm is achieved on test specimens (mouse and amphibian meiotic chromosomes). Finally, stimulated emission depletion (STED) microscopy and LFSM are demonstrated on the same biological sample using a simple commercial mounting medium. It is hoped that this type of correlative imaging will provide a basis for a further enhanced resolution. 
Dr Kirti Prakash, The Institute of Cancer Research, UK.

Dr Kirti Prakash, The Institute of Cancer Research, UK.Kirti Prakash is a computer scientist by training (Bachelors and Masters degree) but a biologist at heart (PhD degree). Kirti aspires to be an inventor and develop new imaging tools for cell biology and neuroscience. Kirti did his Masters in Computer Science from Aalto University (Finland) and PhD in Biology from Heidelberg University (Germany). During his PhD, he developed a new method to image DNA which led to the first high-resolution images of the epigenetic landscape of meiotic chromosomes and mechanisms behind chromosome condensation. The doctoral research earned him several awards including Springer Best PhD Thesis Prize. After his PhD, he did a couple of postdocs at Carnegie Institution for Science (USA) and University of Cambridge (UK). The primary highlights of his research here were laser-free superresolution microscopy and the development of a high-content imaging pipeline to quantify single-cell gene expression. He then spent a couple of years at the National Physical Laboratory (UK) to work on standardisation and quality control of quantitative microscopy. Presently at the Institute of Cancer Research (UK), he is working on artificial intelligence and digital pathology. |
| 15:50 - 16:10 |
Enabling single-molecule localisation microscopy in turbid food emulsions
Single-molecule detection schemes offer powerful means to overcome static and dynamic heterogeneity. The number of accessible microscopy frameworks that are suitable for dim samples or measurements in turbid food systems, however, has remained low. The researchers therefore developed the miCube: a versatile super-resolution capable microscope, which combines high spatiotemporal resolution, good adaptability, and straight forward installation. They further enabled ultrafast data analysis using a phasor-based localisation algorithm. In the second part, Dr Hohlbein will present results on studying food-related emulsions using super-resolution microscopy. To mitigate the issue of turbidity and to increase the accessible optical resolution in food microscopy, the researchers employed adaptive optics (AO) to compensate aberrations and to modulate the emission wavefront enabling point spread function (PSF) engineering. As a model system for a non-transparent food colloid, they designed an oil-in-water emulsion containing the ferric ion binding protein phosvitin commonly present in egg yolk. They targeted phosvitin with fluorescently labelled primary antibodies and obtained two- and three-dimensional images of phosvitin covered oil droplets. Their data indicated that phosvitin is homogeneously distributed at the interface. With the possibility to obtain super-resolved images in depth, their work paves the way for localising biomacromolecules at heterogeneous colloidal interfaces in food emulsions. 
Dr Johannes Hohlbein, Wageningen University & Research, the Netherlands

Dr Johannes Hohlbein, Wageningen University & Research, the NetherlandsDr Johannes Hohlbein studied Medical Physics at the MLU Halle-Wittenberg (Germany). In 2008 he obtained his PhD in Physics working on single-molecule detection in nanoscale confinement at the MPI of Microstructure Physics (Halle, Germany). He then joined the ‘Gene Machines’ group of Professor Kapanidis at the University of Oxford (UK) to work on DNA polymerases and developing assays for DNA sequencing. In 2012 he accepted a position as an Assistant Professor in the Laboratory of Biophysics at Wageningen University & Research (The Netherlands). Since then, his lab has been working on studying DNA-protein interactions in vitro and advancing single-molecule detection schemes. In 2018, Dr Hohlbein obtained tenure followed by promotion to Associate Professor. The current research in the lab is focussed on exploring DNA-RNA-protein interactions in live bacteria and performing functional food imaging using super-resolution microscopy and related techniques. |
| 16:10 - 16:30 |
Open microscopy documentation for the real world: interactive tools for quality, reproducibility and sharing value of imaging experiments based on community specifications
For quality, interpretation, reproducibility and sharing value, microscopy images should be accompanied by detailed descriptions of the conditions that were used to produce them. In this talk Dr Strambio De Castillia will discuss highly interoperable, open-source software tools that were designed in the context of burgeoning global bioimaging community initiatives to facilitate the documentation of microscopy experiments as specified by the recent 4DN-BINA-OME tiered-system of Microscopy Metadata specifications. In addition to substantially lowering the burden of quality assurance, these tools visual nature of these tools make them well suited for teaching users about the intricacies of image acquisition and how it impacts the results of their experiments. 
Dr Caterina Strambio De Castillia, The University of Massachusetts Chan Medical School, USA

Dr Caterina Strambio De Castillia, The University of Massachusetts Chan Medical School, USADr Strambio De Castillia is a faculty member at UMass Medical School and a Chan Zuckerberg Initiative Imaging Scientist. She has multifaceted training spanning cellular biology, biochemistry, and mass spectrometry, virology, electron and light microscopy, computer science, and bio-image informatics. She has an ongoing interest in elucidating the fundamental principles governing the intracellular trafficking of viruses, the dynamic regulation of nucleocytoplasmic transport as it interfaces with nuclear envelope structure and with chromatin organisation and function. More recently, she spearheaded several multi-disciplinary bio-image informatics projects and initiatives aimed at promoting scientific rigor, reproducibility, and the FAIR exchange of data through the standardisation of metadata guidelines and quality-control procedures, and the development of shared software infrastructure for imaging pipelines. |
| 16:30 - 16:45 |
clesperanto: Open-source GPU-accelerated image processing across programming languages and software ecosystems
The optimised computing power of graphics processing units (GPUs) is changing the way we do image analysis in the life sciences. Not just new deep learning approaches but also GPU-accelerated classical image processing techniques are becoming available to end-users with minimal coding skills. This ongoing revolution is an opportunity to synchronise image processing operations and workflows as many of them have to be rewritten for new GPU-based computing architectures. The clesperanto project is paving the path for image-analysts working with Fiji, Icy, ImageJ, Matlab, napari, Jython, Python and others to use the same language for formulating their scientific image analysis workflows in a cross-platform fashion and thus, connects the communities of multiple software ecosystems. 
Dr Robert Haase, University of Technology TU Dresden, Germany

Dr Robert Haase, University of Technology TU Dresden, GermanyDr Haase graduated in computer science at the University of Applied Sciences Dresden and did a PhD in medical image processing at the Medical Faculty Carl Gustav Carus at the University of Technology TU Dresden, Germany. After deepening his expertise in bio-image analysis in the microscopy context in the Scientific Computing Facility and in Gene Myers lab at the Max Planck Institute for Molecular Cell Biology and Genetics in Dresden, he became group leader for Bio-image Analysis Technology Development at the DFG Cluster of Excellence 'Physics of Life' at the TU Dresden. His group is affiliated with and located at the Center for Systems Biology in Dresden. His research focuses on leveraging high-performance computing and facilitating advanced bio-image analysis and image data science techniques in the life sciences. |
| 16:45 - 17:45 |
Panel discussion - Open Microscopy: Quo Vadis?

Dr Johannes Hohlbein, Wageningen University & Research, the Netherlands

Dr Johannes Hohlbein, Wageningen University & Research, the NetherlandsDr Johannes Hohlbein studied Medical Physics at the MLU Halle-Wittenberg (Germany). In 2008 he obtained his PhD in Physics working on single-molecule detection in nanoscale confinement at the MPI of Microstructure Physics (Halle, Germany). He then joined the ‘Gene Machines’ group of Professor Kapanidis at the University of Oxford (UK) to work on DNA polymerases and developing assays for DNA sequencing. In 2012 he accepted a position as an Assistant Professor in the Laboratory of Biophysics at Wageningen University & Research (The Netherlands). Since then, his lab has been working on studying DNA-protein interactions in vitro and advancing single-molecule detection schemes. In 2018, Dr Hohlbein obtained tenure followed by promotion to Associate Professor. The current research in the lab is focussed on exploring DNA-RNA-protein interactions in live bacteria and performing functional food imaging using super-resolution microscopy and related techniques. 
Dr Benedict Diederich, Leibniz Institute of Photonic Technology, Germany

Dr Benedict Diederich, Leibniz Institute of Photonic Technology, GermanyAfter doing an apprenticeship as an electrician, Benedict Diederich started studying electrical engineering at the University for Applied Science Cologne. A specialization in optics and an internship at Nikon Microscopy Japan pointed him to the interdisciplinary field of microscopy. After working for Zeiss, he started his PhD in the Heintzmann Lab at the Leibniz IPHT Jena. He focuses on bringing cutting-edge research to everybody by relying on tailored image processing and low-cost optical setups. Part of his PhD program took place at the Photonics Centre at Boston University in the Tian Lab. A recent contribution was the open-source optical toolbox UC2 (You-See-Too) which tries to democratize science by making cutting-edge affordable and available to everyone, everywhere. He is co-founder of the open-hardware start-up “openUC2” which aims to scale up open microscopy to solve global problems and spend some time during his Post Doc in Manu Prakash’s lab at Stanford University to promote “Frugal Optics” in marine biology communities. 
Dr Robert Haase, University of Technology TU Dresden, Germany

Dr Robert Haase, University of Technology TU Dresden, GermanyDr Haase graduated in computer science at the University of Applied Sciences Dresden and did a PhD in medical image processing at the Medical Faculty Carl Gustav Carus at the University of Technology TU Dresden, Germany. After deepening his expertise in bio-image analysis in the microscopy context in the Scientific Computing Facility and in Gene Myers lab at the Max Planck Institute for Molecular Cell Biology and Genetics in Dresden, he became group leader for Bio-image Analysis Technology Development at the DFG Cluster of Excellence 'Physics of Life' at the TU Dresden. His group is affiliated with and located at the Center for Systems Biology in Dresden. His research focuses on leveraging high-performance computing and facilitating advanced bio-image analysis and image data science techniques in the life sciences. 
Dr Caterina Strambio De Castillia, The University of Massachusetts Chan Medical School, USA

Dr Caterina Strambio De Castillia, The University of Massachusetts Chan Medical School, USADr Strambio De Castillia is a faculty member at UMass Medical School and a Chan Zuckerberg Initiative Imaging Scientist. She has multifaceted training spanning cellular biology, biochemistry, and mass spectrometry, virology, electron and light microscopy, computer science, and bio-image informatics. She has an ongoing interest in elucidating the fundamental principles governing the intracellular trafficking of viruses, the dynamic regulation of nucleocytoplasmic transport as it interfaces with nuclear envelope structure and with chromatin organisation and function. More recently, she spearheaded several multi-disciplinary bio-image informatics projects and initiatives aimed at promoting scientific rigor, reproducibility, and the FAIR exchange of data through the standardisation of metadata guidelines and quality-control procedures, and the development of shared software infrastructure for imaging pipelines. 
Dr Stefanie Reichelt, University of Cambridge, UK

Dr Stefanie Reichelt, University of Cambridge, UKStefanie established and led the light microscopy core at the Cancer Research UK Cambridge Institute (CRUK CI) for 16 years, which included the development of new imaging techniques enabling the visualisation of molecules in cells for cancer diagnostics. Prior to this, Stefanie contributed to the development and commercialisation of laser scanning microscopy with Brad Amos, FRS, at the MRC-LMB and Bio-Rad Microscience. She is the co-founder of the Plymouth Advanced Microscopy Course and has established the Cambridge Technology Platforms Network (CTPN) and the Royal Microscopical Society Imaging One World lecture series. In her current role as Public Engagement Manager for the Biomedical schools at the University of Cambridge, UK, Stefanie contributes to the public understanding of science. |
| 17:45 - 17:55 | Closing remarks |
