BioStudies is a database that aims to link research articles with all the data behind a study. Here, Johanna McEntyre and Ugis Sarkans, based at the European Molecular Biology Laboratory, European Bioinformatics Institute (EMBL-EBI), Cambridge, UK tell us a bit more about it.

BioStudies is a database that aims to link research articles with all the data behind a study. Here, Johanna McEntyre and Ugis Sarkans, based at the European Molecular Biology Laboratory, European Bioinformatics Institute (EMBL-EBI), Cambridge, UK tell us a bit more about it.
Biological experiments produce abundant and heterogeneous data, yet the primary and most important means of communicating results remains the research paper, the format of which has changed little over the past 50 years. It is a major challenge to integrate modern research outputs, in this case data, to this tried and tested format. While we are not lacking technical solutions, driving uptake is much harder, as established workflows need to be rerouted and behaviour from not only researchers but also publishers, funders and institutions adjusts. The goal of the BioStudies database is to support this change by providing a simple solution to linking research articles with all the data behind a study, contributing to the reproducibility of research and opening possibilities for deeper integration of research objects in the future. Launched in 2015, BioStudies is a relatively new database, developed at the European Bioinformatics Institute (EMBL-EBI), and is increasingly being used by a number of projects as well as individual researchers.
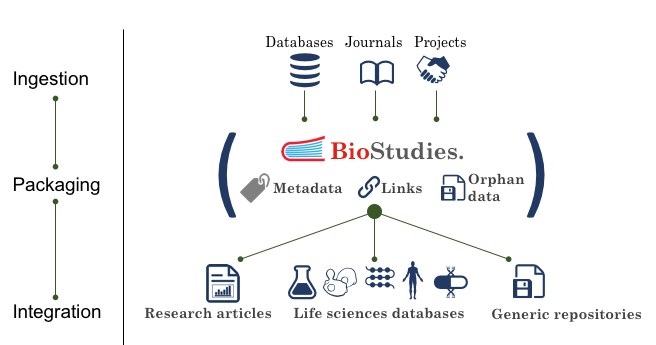
In the life sciences, most major types of data have international community deposition databases in which data should be put to make it Findable, Accessible, Interoperable, and Reusable (FAIR). These data resources typically operate according to international standards of data formats and metadata collection and are often mentioned in papers as Accession numbers, many of which are instantly recognisable by biologists as belonging to the Protein Data Bank, PDB (6NFT), European Nucleotide Archive (KY426898) or Ensembl (ENSG00000140379), to name a few. Other data outputs are often attached to a paper as supplemental data files, or, less frequently, deposited in generic data repositories such as Figshare, Zenodo, or Dryad or in an Institutional Repository, and cited in the paper as a DOI. Given the diversity of locations in which data behind a study may be found, and the high variability in quality of linking data with papers across thousands of journals, it becomes increasingly difficult over time to find all the data that support the science reported in a paper.
BioStudies packages all the data about a study. Each record contains a description of the study, links to data in community resources and generic resources, and files of other “orphan” data that relates to the structured data in community resources (see figure). This complements, rather than supplants, established practices to improve integration and findability. Furthermore, because all the data is aggregated into one citable record, it makes it easier for authors and journal editors to link effectively to the data behind a paper, for example, from Data Availability sections. A BioStudies record can be cited with either a DOI or Accession number. We hope that making it easier for researchers to manage and cite data will encourage good practice and improve transparency of research in the longer term.
A major user of BioStudies is Europe PMC. All Europe PMC articles that mention data (detected through text mining) and/or have supplemental data files, now have a corresponding BioStudies record – currently just under 1.2 million articles. Simply having BioStudies records linked from Europe PMC records makes it easier to find all the data behind a paper (see, for example, the BioStudies link under the abstract). But BioStudies records are not required to be linked to a paper. This means that you can start to package and publish your data prior to publication, ready for citation, and indeed, make further additions to the record after a paper is published (BioStudies has built-in versioning). While publishing data prior to publication is best practice, many authors may think of depositing supplemental data files only at the time of publication. Therefore we are working with journals to integrate BioStudies into article submission workflows.
“We have been delighted to work with Open Biology as an early adopter of such an integration, a collaboration that provides immediate benefits to data management, but also informs future development plans.”
Further reading:
Sarkans U, Gostev M, Athar A, Behrangi E, Melnichuk O, Ali A, Minguet J, Rada JC, Snow C, Tikhonov A, Brazma A, McEntyre J. The BioStudies database-one stop shop for all data supporting a life sciences study. Nucleic Acids Res. 2018 Jan;4s6(D1) D1266-D1270. doi:10.1093/nar/gkx965. PMID: 29069414; PMCID: PMC5753238.
McEntyre J, Sarkans U, Brazma A. The BioStudies database. Mol Syst Biol. 2015 Dec;11(12) 847. doi:10.15252/msb.20156658. PMID: 26700850; PMCID: PMC4704487.
Open Biology is looking to publish more high quality research articles in cellular and molecular biology. Find out more about our author benefits and submission process.




