New research published in Proceedings B takes a modelling approach to investigate co-infection scenarios of different respiratory viruses.
In these troubling times where the global Covid-19 pandemic rages on, Proceedings B has published a study that investigated a virus–virus interaction between respiratory viruses, Covid-19 and influenza. The study showed that using a common epidemiological study design called the test-negative design generally failed at estimating the strength, and sometimes even the direction of virus-virus interactions. Hence, more studies will be needed to elucidate the epidemiological interactions of SARS-CoV-2 and other respiratory viruses. Lead author Matthieu Domenech de Cellès from the Max Planck Institute for Infection Biology tells us more about the study.
Early in the COVID-19 pandemic, we became interested in the potential interactions of SARS-CoV-2 with other co-circulating pathogens, in particular with influenza viruses. Early experimental studies (first in cell cultures, then in animal models) had indicated that influenza can up-regulate the expression of ACE2 (the cognate receptor of SARS-CoV-2 on human cells) and may thus facilitate secondary infection with SARS-CoV-2. At the population scale, however, the evidence was inconsistent, with some studies reporting synergistic interactions, and others neutral or antagonistic interactions. Upon review, we noted that the latter studies used a common epidemiological design—called the test-negative design—to estimate the difference in the risk of SARS-CoV-2 infection between individuals testing positive or negative to influenza. The underlying idea of this design is conceptually simple and stems from standard statistical tests of independence. Specifically, if influenza and SARS-CoV-2 do not interact and therefore circulate independently, then one expects the frequency of co-detection estimated from cross-sectional data to be approximately equal to the product of each virus’s detection frequency. Although this seems intuitive, we also knew from previous studies that such simple statistics can be misleading measures of interaction. However, evidence specific to respiratory viruses was lacking. This motivated us to investigate further and to conduct a simulation study, because mathematical models are an excellent tool to assess study design via controlled numerical experiments.
The results were both clear-cut and striking: we found that studies based on the test-negative design systematically under-estimated the strength of interaction, and could even misclassify (e.g., conclude that interaction is synergistic when it is in fact antagonistic) interactions that persist after clearance of infection. In essence, the complex, non-linear dynamics of respiratory viruses makes the interpretation of seemingly intuitive measures of interaction difficult, if not impossible.
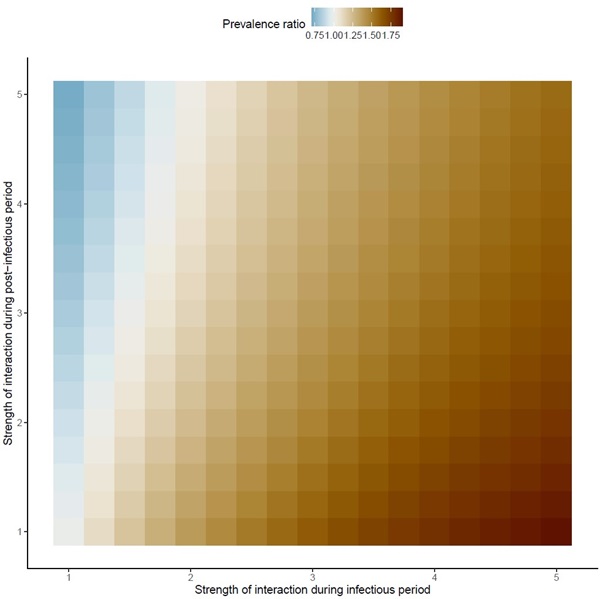
The important point is that one would expect all colors to be red in this figure, because the true interaction was set to positive in these simulations. Hence, the regions in blue represent scenarios in which the test-negative design would have led to the wrong conclusion (that is, that interaction was negative, when in fact it was positive).
About the authors
Matthieu Domenech de Cellès. I head the “Infectious Disease Epidemiology” research group at the Max Planck Institute for Infection Biology in Berlin. Created in January 2020, the group aims to conduct research in the field of infectious disease epidemiology and ecology, using a variety of mathematical and statistical modeling tools. An axis of research in the lab—from which this paper originates—focuses on pathogen interactions, and more specifically on interactions between seasonal (e.g., influenza viruses, RSV, rhinoviruses, etc.) or emerging (e.g., SARS-CoV-2) respiratory viruses. In addition to being scientifically exciting, such interactions may also be consequential from a public health perspective. Indeed, the existence of interaction implies that interventions (for example vaccination) against a given pathogen may have indirect effects against other pathogens—potentially opening new avenues for control measures.
Elizabeth Goult. Having previously completed her masters in mathematical modelling at University College London, Elizabeth Goult began her academic career modelling ocean ecosystems, before moving to modelling the dynamics of cholera at Plymouth Marine Laboratory. She is now a PhD student at the Max Planck Institute for Infection Biology, supervised by Dr Domenech de Cellès, interested in understanding the seasonality of viral respiratory infections.
Dr. Jean-Sébastien Casalegno is a medical doctor and virologist, interested in respiratory viruses. He published some of the earliest studies on epidemiological interference between respiratory viruses during the 2009 A(H1N1) pandemic.
Dr. Sarah Kramer is a postdoctoral researcher at the Max Planck Institute for Infection Biology in Berlin, Germany. Her research uses mathematical and statistical modeling to better understand infectious disease transmission dynamics, including the impact of seasonality and virus-virus interactions. Previously, Sarah obtained her PhD in Environmental Health Sciences from the Columbia University Mailman School of Public Health, where she worked on forecasting influenza outbreaks.

From right to left: Elizabeth Goult, Sarah Kramer, and Matthieu (Anabelle Wong and Laura Barrero, on the left, did not participate in this study).
What was your experience like publishing in Proceedings B?
Overall, the experience was great, and we are grateful to the associate editor for handling our manuscript quickly and effectively. Our two reviewers were also excellent, and their comments helped improve and increase the scope of the study. I had similarly positive experience for two other papers previously published in Proceedings B, and more generally I greatly enjoy reading research in this journal.
Proceedings B is looking to publish more high-quality research articles and reviews in health, disease and epidemiology. If you have an idea for a review, we strongly encourage you to submit a proposal by completing our proposal template and sending it to the journal. More information about the journal and the submission process can be found on our website.
Image credits:
2. Adapted from Figure 3. The pitfalls of inferring virus-virus interactions from codetection prevalence data: Application to influenza and SARS-CoV-2
3. Photo by David Ausserhofer © Max Planck Institute for Infection Biology.




