To celebrate Open Access Week, we highlight some of our best-performing articles from the past year. We hope you’ll enjoy reading them as much as we have!
.jpg)
Plant NLR immunity activation and execution: a biochemical perspective
Did you know that plants have their own defence systems to fight off diseases? They use a wide range of receptor proteins like nucleotide-binding/leucine-rich repeat (NLR) proteins to detect attacks from harmful microbes, and when these receptors sense a pathogen, they activate defence mechanisms that stop the disease in its tracks. Recent advances in biochemistry have shed light on how these receptors get activated and signal the plant to resist disease. Once triggered, the receptors cause certain small molecules to build up, acting as messengers that turn on the plant's defence programs within its cells and tissues. By recognising invading pathogens, these receptors use these "stress" signals to initiate a broad immune response (including changes in cellular calcium levels) that prevent the pathogens from growing and spreading in the plant.
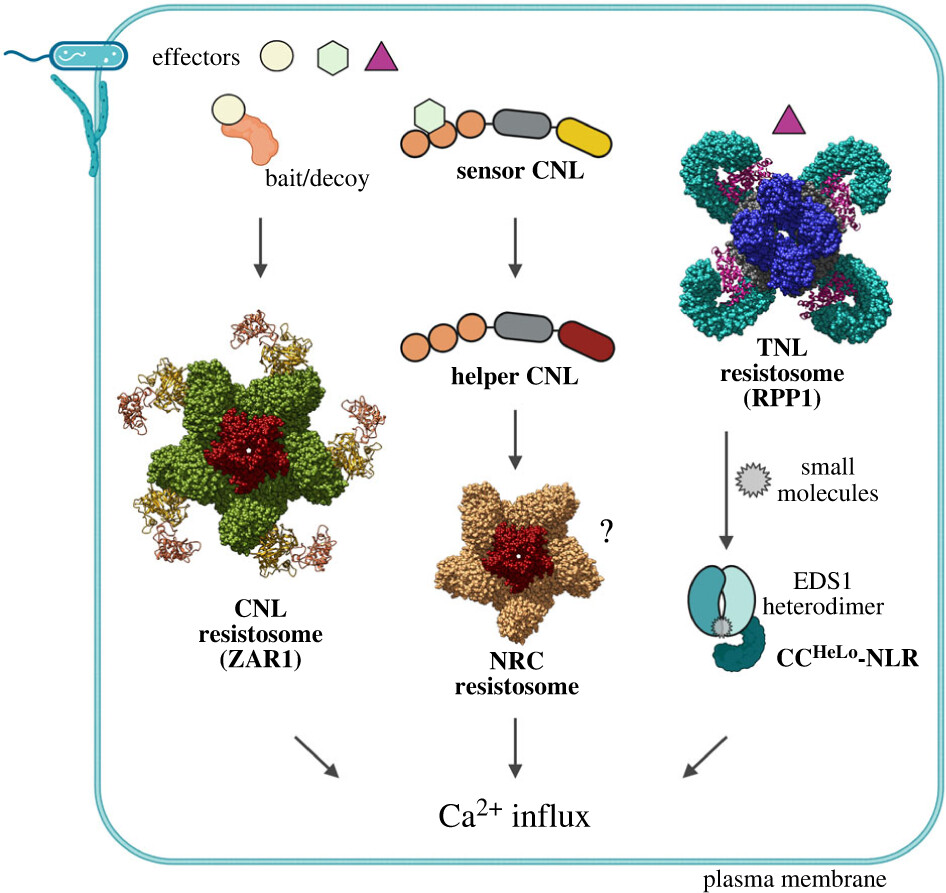
Different plant NLR resistosomes converge on Ca2+ influx in immunity (doi.org/10.1098/rsob.230387).
Breakthrough therapy could halt most respiratory viruses early
Respiratory illnesses are notoriously difficult to treat because they're caused by a variety of viruses, including those that trigger asthma and COPD flare-ups, and effective treatment must start before the exact virus is identified. Additionally, viruses often develop resistance to drugs and vaccines that target them directly. While many respiratory infections are mild, some become serious and nearly all begin in the upper respiratory tract. A new drug works against most human respiratory viruses by targeting a novel interaction point between the virus and our cells - a spot where viruses can't develop resistance. This means it could be used at the first sign of symptoms, like a sore throat or runny nose, to prevent the illness from progressing to more severe lower respiratory tract disease.
Exploring the role of risk gene CNTN4 and APP in neuronal development
This paper explored the intricate relationship between the neuronal cell adhesion molecule contactin-4 (CNTN4) and amyloid precursor protein (APP) in neurodevelopmental disorders and Alzheimer's disease. The study reveals that CNTN4 is crucial in shaping neuronal morphology and spine density, and it interacts co-dependently with APP to promote neurite outgrowth, unveiling a novel compensatory expression mechanism between these proteins. Motivated by the limited understanding of CNTN4's functional roles, especially its link to autism spectrum disorders in 3p26 deletion syndrome, the researchers investigated its brain function and interactions with proteins involved in neurodegenerative diseases. “It was quite remarkable to discover that CNTN4, a gene linked to developmental processes, also plays a role in modulating factors involved in Alzheimer's disease. This intersection of developmental and neurodegenerative pathways offers exciting new insights into the broader implications of these proteins.” - Author, Asami Oguro-Ando, University of Exeter.

Co-authors Madeline Eve and Asami Oguro-Ando, University of Exeter.
Amyloidogenic sequence regions control the toxicity and aggregation of mutant SOD1
Researchers have made a significant breakthrough in understanding amyotrophic lateral sclerosis (ALS), a devastating disease linked to mutations in the SOD1 protein. Despite the variety of mutation types and locations within SOD1, current research points to a common toxic element was at play. Utilising advanced computational models and cellular biology, the authors of this study identified two specific regions of the SOD1 protein that contribute to its toxicity and the progression of ALS. This discovery opens the door to developing targeted treatments aimed at these toxic regions, potentially reducing the harmful effects of the disease in patients.
The weird and wonderful world of Monotremes: unique, inside and out
Researchers at the University of Adelaide uncovered a genetic key to understanding the unusual stomach anatomy of platypuses and echidnas. A gene called Nkx3.2, which became inactive in a common ancestor of these monotremes millions of years ago, likely contributed to their small, non-acidic stomachs and, in platypuses, the absence of a pyloric sphincter. The team discovered that Nkx3.2 wasn't functional in monotremes, shedding light on how their distinctive stomachs evolved. "Work from our lab previously had shown that the platypus and echidna had lost the genetic instructions for proteins that break down food and secrete stomach acids, but to me this didn't explain the drastic shift in their stomach anatomy relative to other animals," - first author Jackson Dann, University of Adelaide. The findings provide new insights into the evolution of monotremes, the world's oldest living mammals.

Echidnas belong to the family Tachyglossidae in the monotreme order of egg-laying mammals. Credit: @PatWeeSit Flickr CC-BY 2.0.
Open Biology is seeking high-quality research articles across cellular and molecular biology. Find out more about our author benefits and submission process. Main image: Baby Erinaceus europaeus. Credit: Enrico Cian, Wikimedia Commons.
